9IUK
 
 | | The structure of Candida albicans Cdr1 in apo state | | Descriptor: | Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-22 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9IUM
 
 | |
7XTU
 
 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTR
 
 | | The apo structure of the engineered TfCut | | Descriptor: | GLYCEROL, LITHIUM ION, SULFATE ION, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
9IUL
 
 | | The structure of Candida albicans Cdr1 in fluconazole-bound state | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Pip2(20:4/18:0), Pleiotropic ABC efflux transporter of multiple drugs CDR1 | | Authors: | Peng, Y, Sun, H, Yan, Z.F. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Candida albicans Cdr1 reveal azole-substrate recognition and inhibitor blocking mechanisms.
Nat Commun, 15, 2024
|
|
9KBT
 
 | |
9KBQ
 
 | |
9KBS
 
 | |
7MRW
 
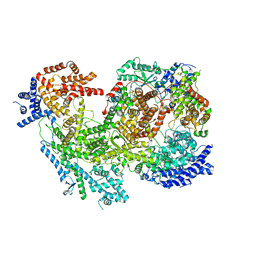 | | Native RhopH complex of the malaria parasite Plasmodium falciparum | | Descriptor: | Cytoadherence linked asexual protein 3.1, High molecular weight rhoptry protein 2, High molecular weight rhoptry protein 3 | | Authors: | Ho, C.M, Jih, J, Lai, M, Li, X.R, Goldberg, D.E, Beck, J.R, Zhou, Z.H. | | Deposit date: | 2021-05-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Native structure of the RhopH complex, a key determinant of malaria parasite nutrient acquisition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8X43
 
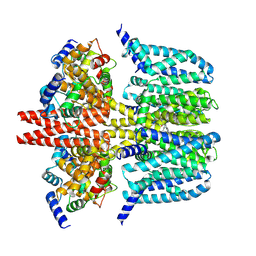 | | human KCNQ2-CaM-Ebio1-S1 complex in the presence of PIP2 | | Descriptor: | Calmodulin-1, N-(4-azanyl-1,2-dihydroacenaphthylen-5-yl)-4-fluoranyl-benzamide, Potassium voltage-gated channel subfamily KQT member 2 | | Authors: | Ma, D, Guo, J. | | Deposit date: | 2023-11-15 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A small-molecule activation mechanism that directly opens the KCNQ2 channel.
Nat.Chem.Biol., 20, 2024
|
|
8YGG
 
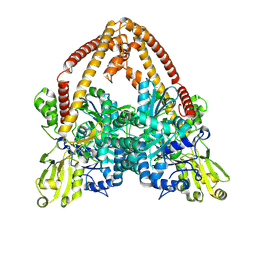 | | pP1192R-apo Closed state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGH
 
 | | pP1192R-apo open state | | Descriptor: | DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YGE
 
 | | pP1192R-DNA-m-AMSA complex DNA binding/cleavage domain | | Descriptor: | DNA (12-mer), DNA (20-mer), DNA (8-mer), ... | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-26 | | Release date: | 2024-09-18 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
8YIK
 
 | | pP1192R-ATPase-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA topoisomerase 2 | | Authors: | Sun, J.Q, Liu, R.L. | | Deposit date: | 2024-02-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for difunctional mechanism of m-AMSA against African swine fever virus pP1192R.
Nucleic Acids Res., 52, 2024
|
|
7BTK
 
 | | E.coli beta-galactosidase (E537Q) in complex with fluorescent probe KSA01 | | Descriptor: | 4-[[2-[(E)-2-[4-[(2S,3R,4S,5R,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxyphenyl]ethenyl]-3,3-dimethyl-2H-indol-1-yl]methyl]benzoic acid, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Chen, X, Hu, Y.L, Liu, Q.M, Gao, Y, Yuan, R, Guo, Y. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Two-Dimensional Design Strategy to Construct Smart Fluorescent Probes for the Precise Tracking of Senescence.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7KGB
 
 | |
8ZOV
 
 | |
8ZO3
 
 | |
7MT2
 
 | | Mtb 70S initiation complex | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MSM
 
 | | Mtb 70SIC in complex with MtbEttA at Trans_R0 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
7MSZ
 
 | | Mtb 70SIC in complex with MtbEttA at Trans_R1 state | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2021-05-12 | | Release date: | 2022-02-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Interplay between an ATP-binding cassette F protein and the ribosome from Mycobacterium tuberculosis.
Nat Commun, 13, 2022
|
|
9K3Z
 
 | |
9K40
 
 | |
9K42
 
 | |
9KRQ
 
 | | Crystal structure of ZXC21 RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lan, J, Wang, C.H. | | Deposit date: | 2024-11-28 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | Structural insights into the receptor-binding domain of bat coronavirus ZXC21.
Structure, 2025
|
|
