2MAW
 
 | |
1VRY
 
 | | Second and Third Transmembrane Domains of the Alpha-1 Subunit of Human Glycine Receptor | | Descriptor: | Glycine receptor alpha-1 chain | | Authors: | Ma, D, Liu, Z, Li, L, Tang, P, Xu, Y. | | Deposit date: | 2005-07-20 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the Second and Third Transmembrane Domains of Human Glycine Receptor.
Biochemistry, 44, 2005
|
|
4F8H
 
 | | X-ray Structure of the Anesthetic Ketamine Bound to the GLIC Pentameric Ligand-gated Ion Channel | | Descriptor: | (R)-ketamine, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Proton-gated ion channel, ... | | Authors: | Pan, J.J, Chen, Q, Willenbring, D, Kong, X.P, Cohen, A, Xu, Y, Tang, P. | | Deposit date: | 2012-05-17 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Pentameric Ligand-Gated Ion Channel GLIC Bound with Anesthetic Ketamine.
Structure, 20, 2012
|
|
6AQF
 
 | | Crystal structure of A2AAR-BRIL in complex with the antagonist ZM241385 produced from Pichia pastoris | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Eddy, M.T, Lee, M.Y, Gao, Z, White, K, Didenko, T, Horst, R, Audet, M, Stanczak, P, McClary, K.M, Han, G.W, Jacobson, K.A, Stevens, R.C, Wuthrich, K. | | Deposit date: | 2017-08-19 | | Release date: | 2018-01-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Allosteric Coupling of Drug Binding and Intracellular Signaling in the A2A Adenosine Receptor.
Cell, 172, 2018
|
|
2BRN
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | (2R)-1-[(5,6-DIPHENYL-7H-PYRROLO[2,3-D]PYRIMIDIN-4-YL)AMINO]PROPAN-2-OL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-09 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2C3L
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOLE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2BRH
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | N-(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)GLYCINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-05 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2C3K
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | 4-[3-(1H-BENZIMIDAZOL-2-YL)-1H-INDAZOL-6-YL]-2-METHOXYPHENOL, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2BRB
 
 | | Structure-based Design of Novel Chk1 Inhibitors: Insights into Hydrogen Bonding and Protein-Ligand Affinity | | Descriptor: | 2-[(5,6-DIPHENYLFURO[2,3-D]PYRIMIDIN-4-YL)AMINO]ETHANOL, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFATE ION | | Authors: | Foloppe, N, Fisher, L.M, Howes, R, Kierstan, P, Potter, A, Robertson, A.G.S, Surgenor, A.E. | | Deposit date: | 2005-05-04 | | Release date: | 2005-05-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of Novel Chk1 Inhibitors: Insights Into Hydrogen Bonding and Protein-Ligand Affinity.
J.Med.Chem., 48, 2005
|
|
2C3J
 
 | | Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification | | Descriptor: | DEBROMOHYMENIALDISINE, SERINE/THREONINE-PROTEIN KINASE CHK1, SULFITE ION | | Authors: | Foloppe, N, Fisher, L.M, Francis, G, Howes, R, Kierstan, P, Potter, A. | | Deposit date: | 2005-10-10 | | Release date: | 2005-11-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a Buried Pocket for Potent and Selective Inhibition of Chk1: Prediction and Verification.
Bioorg.Med.Chem., 14, 2006
|
|
2Y1X
 
 | | CRYSTAL STRUCTURE OF COACTIVATOR ASSOCIATED ARGININE METHYLTRANSFERASE 1 (CARM1) IN COMPLEX WITH SINEFUNGIN AND INDOLE INHIBITOR | | Descriptor: | CHLORIDE ION, HISTONE-ARGININE METHYLTRANSFERASE CARM1, N-(3-{5-[5-(1H-INDOL-4-YL)-1,3,4-OXADIAZOL-2-YL]-3-(TRIFLUOROMETHYL)-1H-PYRAZOL-1-YL}BENZYL)-L-ALANINAMIDE, ... | | Authors: | Sack, J.S, Thieffine, S, Bandiera, T, Fasolini, M, Duke, G.J, Jayaraman, L, Kish, K.F, Klei, H.E, Purandare, A.V, Rosettani, P, Troiani, S, Xie, D, Bertrand, J.A. | | Deposit date: | 2010-12-10 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis for Carm1 Inhibition by Indole and Pyrazole Inhibitors
Biochem.J., 436, 2011
|
|
3XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, sorbitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
3ZUR
 
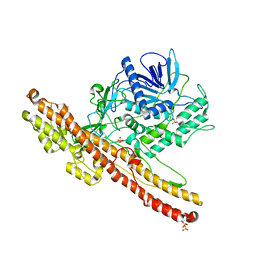 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC0-A-SNAP25-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN, SULFATE ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZUQ
 
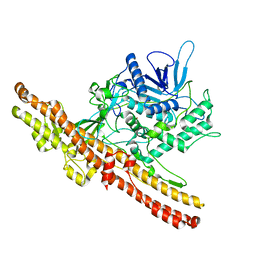 | | Crystal structure of an engineered botulinum neurotoxin type B - derivative, LC-B-GS-Hn-B | | Descriptor: | BOTULINUM NEUROTOXIN TYPE B, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3ZH9
 
 | | Bacillus subtilis DNA clamp loader delta protein (YqeN) | | Descriptor: | DELTA, GLYCEROL, SULFATE ION | | Authors: | Suwannachart, C, Sedelnikova, S, Soultanas, P, Oldham, N.J, Rafferty, J.B. | | Deposit date: | 2012-12-20 | | Release date: | 2013-04-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insights Into the Structure and Assembly of the Bacillus Subtilis Clamp-Loader Complex and its Interaction with the Replicative Helicase.
Nucleic Acids Res., 41, 2013
|
|
3ZUS
 
 | | Crystal structure of an engineered botulinum neurotoxin type A- SNARE23 derivative, LC-A-SNAP23-Hn-A | | Descriptor: | BOTULINUM NEUROTOXIN TYPE A, SYNAPTOSOMAL-ASSOCIATED PROTEIN 23, ZINC ION | | Authors: | Masuyer, G, Stancombe, P, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2011-07-19 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of Engineered Clostridium Botulinum Neurotoxin Derivatives
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4AXK
 
 | | CRYSTAL STRUCTURE OF subHisA from the thermophile Corynebacterium efficiens | | Descriptor: | 1-(5-PHOSPHORIBOSYL)-5-((5'-PHOSPHORIBOSYLAMINO) METHYLIDENEAMINO)IMIDAZOLE-4-CARBOXAMIDE ISOMERASE, GLYCEROL | | Authors: | Noda-Garcia, L, Camacho-Zarco, A.R, Medina-Ruiz, S, Verduzco-Castro, E.A, Gaytan, P, Carrillo-Tripp, M, Fulop, V, Barona-Gomez, F. | | Deposit date: | 2012-06-13 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of Substrate Specificity in a Recipient'S Enzyme Following Horizontal Gene Transfer.
Mol.Biol.Evol., 30, 2013
|
|
8CH5
 
 | |
9XIM
 
 | |
7NWR
 
 | | Structure of BT1526, a myo-inositol-1-phosphate synthase | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Basle, A, Tang, G, Marles-Wright, J, Campopiano, D. | | Deposit date: | 2021-03-17 | | Release date: | 2022-03-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of inositol lipid metabolism in gut-associated Bacteroidetes.
Nat Microbiol, 7, 2022
|
|
8AHH
 
 | |
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHG
 
 | |
8AHI
 
 | |
