3EMY
 
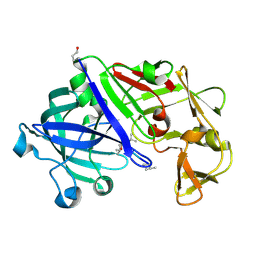 | | Crystal structure of Trichoderma reesei aspartic proteinase complexed with pepstatin A | | Descriptor: | Pepstatin, Trichoderma reesei Aspartic protease | | Authors: | Nascimento, A.S, Krauchenco, S, Golubev, A.M, Gustchina, A, Wlodawer, A, Polikarpov, I. | | Deposit date: | 2008-09-25 | | Release date: | 2008-10-07 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Statistical coupling analysis of aspartic proteinases based on crystal
structures of the Trichoderma reesei enzyme and its complex with pepstatin A.
J.Mol.Biol., 382, 2008
|
|
3GWS
 
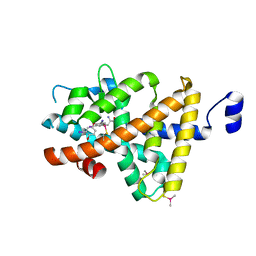 | | Crystal Structure of T3-Bound Thyroid Hormone Receptor | | Descriptor: | 3,5,3'TRIIODOTHYRONINE, Thyroid hormone receptor beta | | Authors: | Nascimento, A.S, Dias, S.M.G, Nunes, F.M, Aparicio, R, Polikarpov, I, Baxter, J.D, Webb, P. | | Deposit date: | 2009-04-01 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rearrangements in the thyroid hormone receptor hinge domain and their putative role in the receptor function
J.Mol.Biol., 360, 2006
|
|
3EXD
 
 | |
3HDL
 
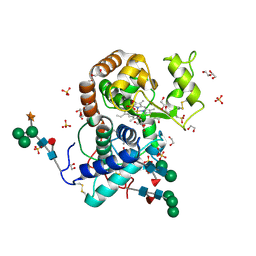 | | Crystal Structure of Highly Glycosylated Peroxidase from Royal Palm Tree | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Watanabe, L, Moura, P.R, Bleicher, L, Nascimento, A.S, Zamorano, L.S, Calvete, J.J, Bursakov, S, Roig, M.G, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2009-05-07 | | Release date: | 2009-11-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and statistical coupling analysis of highly glycosylated peroxidase from royal palm tree (Roystonea regia).
J.Struct.Biol., 169, 2010
|
|
4EMA
 
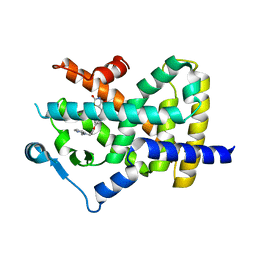 | | Human peroxisome proliferator-activated receptor gamma in complex with rosiglitazone | | Descriptor: | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL), Peroxisome proliferator-activated receptor gamma | | Authors: | Liberato, M.V, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.545 Å) | | Cite: | Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) Gamma activators and pan-PPAR partial agonists
Plos One, 7, 2012
|
|
5J7N
 
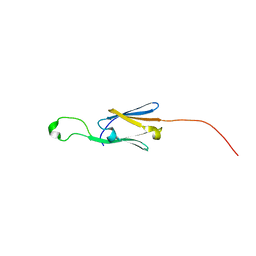 | | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high order structure | | Descriptor: | Low molecular weight heat shock protein | | Authors: | Fonseca, E.M.B, Scorsato, V, dos Santos, C.A, Tomazini Jr, A, Aparicio, R, Polikarpov, I. | | Deposit date: | 2016-04-06 | | Release date: | 2017-04-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a small heat-shock protein from Xylella fastidiosa reveals a distinct high-order structure.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5JBO
 
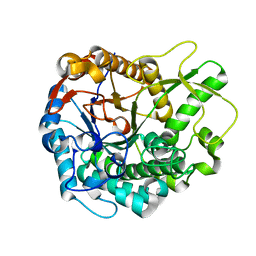 | | Trichoderma harzianum GH1 beta-glucosidase ThBgl2 | | Descriptor: | Beta-glucosidase | | Authors: | Florindo, R.N, Mutti, H.S, Polikarpov, I, Nascimento, A.S. | | Deposit date: | 2016-04-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum.
N Biotechnol, 40, 2018
|
|
4EM9
 
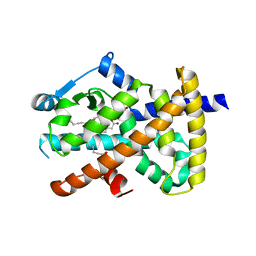 | | Human PPAR gamma in complex with nonanoic acids | | Descriptor: | 3,3',3''-phosphanetriyltripropanoic acid, GLYCEROL, Peroxisome proliferator-activated receptor gamma, ... | | Authors: | Liberato, M.V, Nascimento, A.S, Polikarpov, I. | | Deposit date: | 2012-04-11 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) Gamma activators and pan-PPAR partial agonists
Plos One, 7, 2012
|
|
4DER
 
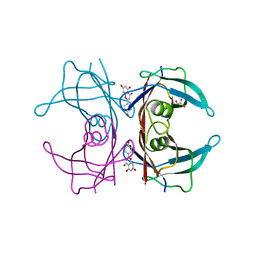 | |
4CVU
 
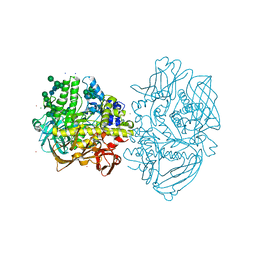 | | Structure of Fungal beta-mannosidase from Glycoside Hydrolase Family 2 of Trichoderma harzianum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-MANNOSIDASE, CADMIUM ION, ... | | Authors: | Muniz, J.R.C, Aparicio, R, Santos, J.C, Nascimento, A.S, Golubev, A.M, Polikarpov, I. | | Deposit date: | 2014-03-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-07-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the structure and function of fungal beta-mannosidases from glycoside hydrolase family 2 based on multiple crystal structures of the Trichoderma harzianum enzyme.
FEBS J., 281, 2014
|
|
4DES
 
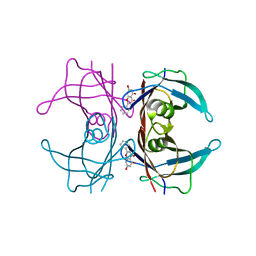 | |
5JBK
 
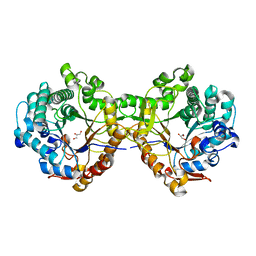 | | Trichoderma harzianum GH1 beta-glucosidase ThBgl1 | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Florindo, R.N, Mutti, H.S, Polikarpov, I, Nascimento, A.S. | | Deposit date: | 2016-04-13 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Structural insights into beta-glucosidase transglycosylation based on biochemical, structural and computational analysis of two GH1 enzymes from Trichoderma harzianum.
N Biotechnol, 40, 2018
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KSQ
 
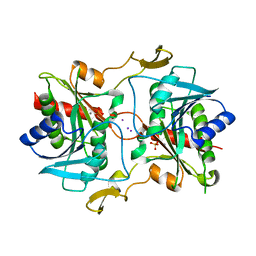 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KJO
 
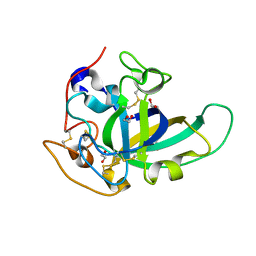 | |
5KJQ
 
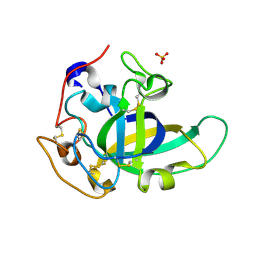 | | X-ray structure of PcCel45A in complex with cellobiose expressed in Aspergillus nidullans | | Descriptor: | Endoglucanase V-like protein, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Godoy, A.S, Ramia, M.P, Camilo, C.M, Polikarpov, I. | | Deposit date: | 2016-06-20 | | Release date: | 2017-06-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Structure, computational and biochemical analysis of PcCel45A endoglucanase from Phanerochaete chrysosporium and catalytic mechanisms of GH45 subfamily C members.
Sci Rep, 8, 2018
|
|
4DET
 
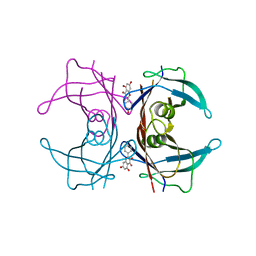 | |
4DEU
 
 | |
4CI5
 
 | |
5KSS
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-Ds (Dimer Smaller) | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, A.M.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5KST
 
 | | Stationary phase Survival protein E (SurE) from Xylella fastidiosa- XfSurE-TSAmp (Tetramer Smaller - crystallization with 3'AMP). | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A.S, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.759 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
4DEW
 
 | |
4CI4
 
 | |
1SZN
 
 | | THE STRUCTURE OF ALPHA-GALACTOSIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brando Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-06 | | Release date: | 2004-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism.
J.Mol.Biol., 339, 2004
|
|
1T0O
 
 | | The structure of alpha-galactosidase from Trichoderma reesei complexed with beta-D-galactose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Golubev, A.M, Nagem, R.A.P, Brandao Neto, J.R, Neustroev, K.N, Eneyskaya, E.V, Kulminskaya, A.A, Shabalin, K.A, Savel'ev, A.N, Polikarpov, I. | | Deposit date: | 2004-04-12 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal structure of alpha-galactosidase from Trichoderma reesei and its complex with galactose: implications for catalytic mechanism
J.Mol.Biol., 339, 2004
|
|
