2MGO
 
 | | NMR solution structure of oxytocin | | Descriptor: | Oxytocin | | Authors: | Harvey, P.J, Craik, D.J. | | Deposit date: | 2013-11-01 | | Release date: | 2014-10-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Oxytocic plant cyclotides as templates for peptide G protein-coupled receptor ligand design.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2M9O
 
 | | Solution structure of kalata B7 | | Descriptor: | Kalata-B7 | | Authors: | Daly, N, Elliott, A, Craik, D. | | Deposit date: | 2013-06-18 | | Release date: | 2013-11-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Cyclotides as templates for peptide GPCR ligand design - discovery of the target receptor of the oxytocic plant peptide kalata B7
Proc.Natl.Acad.Sci.USA, 2013
|
|
7MSA
 
 | | GDC-9545 in complex with estrogen receptor alpha | | Descriptor: | (2S)-3-(3-hydroxyphenyl)-2-(4-iodophenyl)-4-methyl-2H-1-benzopyran-6-ol, 3-[(1R,3R)-1-(2,6-difluoro-4-{[1-(3-fluoropropyl)azetidin-3-yl]amino}phenyl)-3-methyl-1,3,4,9-tetrahydro-2H-pyrido[3,4-b]indol-2-yl]-2,2-difluoropropan-1-ol, Estrogen receptor | | Authors: | Kiefer, J.R, Vinogradova, M, Liang, J, Zbieg, J.R, Wang, X, Ortwine, D.F. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | GDC-9545 (Giredestrant): A Potent and Orally Bioavailable Selective Estrogen Receptor Antagonist and Degrader with an Exceptional Preclinical Profile for ER+ Breast Cancer.
J.Med.Chem., 64, 2021
|
|
5KIT
 
 | |
2KMJ
 
 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
6HPV
 
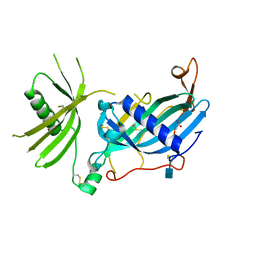 | | Crystal structure of mouse fetuin-B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Fetuin-B | | Authors: | Fahrenkamp, D, Dietzel, E, de Sanctis, D, Jovine, L. | | Deposit date: | 2018-09-22 | | Release date: | 2019-02-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of mammalian plasma fetuin-B and its mechanism of selective metallopeptidase inhibition.
Iucrj, 6, 2019
|
|
6EWU
 
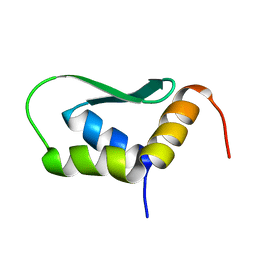 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12C-NDD | | Descriptor: | NRPS Kj12C-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
5AP8
 
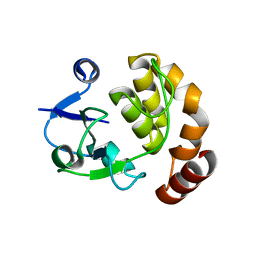 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from S. solfataricus | | Descriptor: | TSR3 | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-14 | | Release date: | 2016-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
6EWT
 
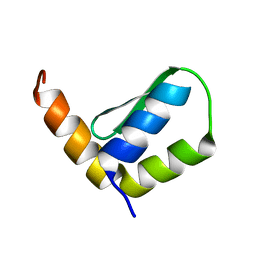 | | Solution Structure of Rhabdopeptide NRPS Docking Domain Kj12B-NDD | | Descriptor: | NRPS Kj12B-NDD | | Authors: | Hacker, C, Cai, X, Kegler, C, Zhao, L, Weickhmann, A.K, Bode, H.B, Woehnert, J. | | Deposit date: | 2017-11-06 | | Release date: | 2018-10-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS.
Nat Commun, 9, 2018
|
|
6WEB
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
6WEC
 
 | | Multi-Hit SFX using MHz XFEL sources | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Holmes, S, Darmanin, C, Abbey, B. | | Deposit date: | 2020-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Megahertz pulse trains enable multi-hit serial femtosecond crystallography experiments at X-ray free electron lasers.
Nat Commun, 13, 2022
|
|
1HMD
 
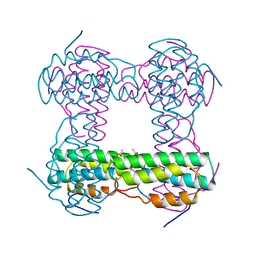 | | THE STRUCTURE OF DEOXY AND OXY HEMERYTHRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL GROUP, HEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Holmes, M.A, Letrong, I, Turley, S, Sieker, L.C, Stenkamp, R.E. | | Deposit date: | 1990-10-18 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of deoxy and oxy hemerythrin at 2.0 A resolution.
J.Mol.Biol., 218, 1991
|
|
1HMO
 
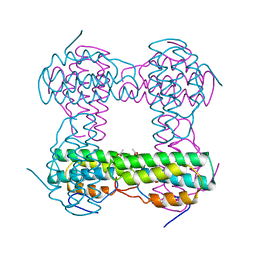 | | THE STRUCTURE OF DEOXY AND OXY HEMERYTHRIN AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | ACETYL GROUP, HEMERYTHRIN, MU-OXO-DIIRON, ... | | Authors: | Holmes, M.A, Letrong, I, Turley, S, Sieker, L.C, Stenkamp, R.E. | | Deposit date: | 1990-10-18 | | Release date: | 1992-01-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of deoxy and oxy hemerythrin at 2.0 A resolution.
J.Mol.Biol., 218, 1991
|
|
2OXI
 
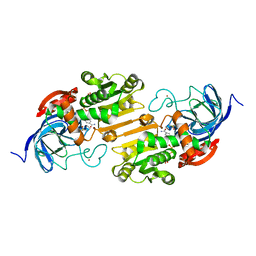 | |
2HMQ
 
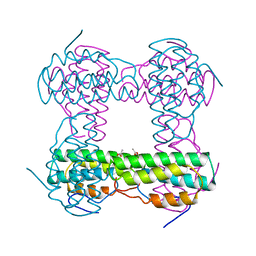 | |
2HMZ
 
 | |
1I4Z
 
 | | THE CRYSTAL STRUCTURE OF PHASCOLOPSIS GOULDII L98Y METHEMERYTHRIN | | Descriptor: | METHEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Farmer, C.S, Kurtz Jr, D.M, Liu, Z.-J, Wang, B.C, Rose, J. | | Deposit date: | 2001-02-23 | | Release date: | 2001-03-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structures of Phascolopsis gouldii wild type and L98Y methemerythrins: structural and functional alterations of the O2 binding pocket.
J.Biol.Inorg.Chem., 6, 2001
|
|
1I4Y
 
 | | THE CRYSTAL STRUCTURE OF PHASCOLOPSIS GOULDII WILD TYPE METHEMERYTHRIN | | Descriptor: | CHLORIDE ION, METHEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Farmer, C.S, Kurtz Jr, D.M, Liu, Z.-J, Wang, B.C, Rose, J. | | Deposit date: | 2001-02-23 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of Phascolopsis gouldii wild type and L98Y methemerythrins: structural and functional alterations of the O2 binding pocket.
J.Biol.Inorg.Chem., 6, 2001
|
|
3ELM
 
 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
7RU4
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.33 IgG heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU1
 
 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU8
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (RBD/Fv local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab Kappa chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU3
 
 | | CC6.33 IgG in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU2
 
 | | SARS-CoV-2-6P-Mut7 S protein (asymmetric) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
7RU5
 
 | | CC6.30 fragment antigen binding in complex with SARS-CoV-2-6P-Mut7 S protein (non-uniform refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CC6.30 Fab heavy chain Fv, ... | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
