4NWI
 
 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, 7-methylguanosine phosphate-specific 5'-nucleotidase, CHLORIDE ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-06 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
4NV0
 
 | | Crystal structure of cytosolic 5'-nucleotidase IIIB (cN-IIIB) bound to 7-methylguanosine | | Descriptor: | 7-METHYLGUANOSINE, 7-methylguanosine phosphate-specific 5'-nucleotidase, MAGNESIUM ION, ... | | Authors: | Monecke, T, Neumann, P, Ficner, R. | | Deposit date: | 2013-12-04 | | Release date: | 2014-03-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Novel Cytosolic 5'-Nucleotidase IIIB Explain Its Preference for m7GMP
Plos One, 9, 2014
|
|
6Z3X
 
 | | Crystal structure of the designed antibody DesAb-anti-HSA-P1 | | Descriptor: | CACODYLATE ION, DesAb-anti-HSA-P1, IMIDAZOLE, ... | | Authors: | Costanzi, E, Sormanni, P, Ricagno, S. | | Deposit date: | 2020-05-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Fragment-based computational design of antibodies targeting structured epitopes.
Sci Adv, 8, 2022
|
|
4NYB
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with (4-(1,2,3-thiadiazol-4-yl)phenyl)methanamine | | Descriptor: | 1-[4-(1,2,3-thiadiazol-4-yl)phenyl]methanamine, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
4NYG
 
 | | Crystal structure of the E. coli thiM riboswitch in complex with thiamine | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, MAGNESIUM ION, thiM TPP riboswitch | | Authors: | Warner, K.D, Homan, P, Weeks, K.M, Smith, A.G, Abell, C, Ferre-D'Amare, A.R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically.
Chem.Biol., 21, 2014
|
|
4O1K
 
 | | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora. | | Descriptor: | Carbonic anhydrase, ZINC ION | | Authors: | Lehneck, R, Neumann, P, Vullo, D, Elleuche, S, Supuran, C.T, Ficner, R, Poggeler, S. | | Deposit date: | 2013-12-16 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structures of two tetrameric beta-carbonic anhydrases from the filamentous ascomycete Sordaria macrospora.
Febs J., 281, 2014
|
|
1UY7
 
 | | Human Hsp90-alpha with 9-Butyl-8-(4-methoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(4-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UYC
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2,5-dimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2,5-DIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
4NOX
 
 | | Structure of the nine-bladed beta-propeller of eIF3b | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit B | | Authors: | Liu, Y, Neumann, P, Kuhle, B, Monecke, T, Ficner, R. | | Deposit date: | 2013-11-20 | | Release date: | 2014-09-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Translation initiation factor eIF3b contains a nine-bladed beta-propeller and interacts with the 40S ribosomal subunit
Structure, 22, 2014
|
|
1UYD
 
 | | Human Hsp90-alpha with 9-Butyl-8-(2-chloro-3,4,5-trimethoxy-benzyl)-9H-purin-6-ylamine | | Descriptor: | 9-BUTYL-8-(2-CHLORO-3,4,5-TRIMETHOXY-BENZYL)-9H-PURIN-6-YLAMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1UY8
 
 | | Human Hsp90-alpha with 9-Butyl-8-(3-trimethoxy-benzyl)-9H-purin-6ylamine | | Descriptor: | 9-BUTYL-8-(3-METHOXYBENZYL)-9H-PURIN-6-AMINE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Wright, L, Barril, X, Dymock, B, Sheridan, L, Surgenor, A, Beswick, M, Drysdale, M, Collier, A, Massey, A, Davies, N, Fink, A, Fromont, C, Aherne, W, Boxall, K, Sharp, S, Workman, P, Hubbard, R.E. | | Deposit date: | 2004-03-02 | | Release date: | 2004-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationships in Purine-Based Inhibitor Binding to Hsp90 Isoforms
Chem.Biol., 11, 2004
|
|
1AGB
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGRKKYKL-3R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGRKKYKL - 3R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGC
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGF
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKRYKL-5R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKRYKL - 5R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGE
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYRL-7R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYRL - 7R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGD
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
4Z3F
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in complex with SSEA4 in space group P21 | | Descriptor: | N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-alpha-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, PapG, lectin domain, ... | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Carbohydrate-Lectin Interactions: An Unexpected Contribution to Affinity.
Chembiochem, 18, 2017
|
|
4Z3J
 
 | | Crystal structure of the lectin domain of PapG from E. coli BI47 in space group P1 | | Descriptor: | PapG, lectin domain | | Authors: | Jakob, R.P, Navarra, G, Zihlmann, P, Stangier, K, Preston, R.C, Rabbani, S, Maier, T, Ernst, B. | | Deposit date: | 2015-03-31 | | Release date: | 2016-04-13 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Carbohydrate-Lectin Interactions: An Unexpected Contribution to Affinity.
Chembiochem, 18, 2017
|
|
7MT0
 
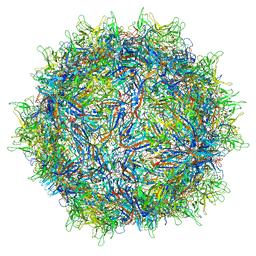 | | Structure of the adeno-associated virus 9 capsid at pH 7.4 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-12 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MUA
 
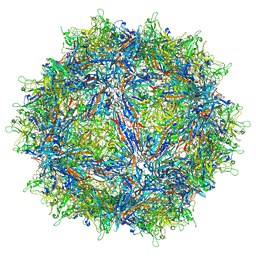 | | Structure of the adeno-associated virus 9 capsid at pH pH 5.5 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTZ
 
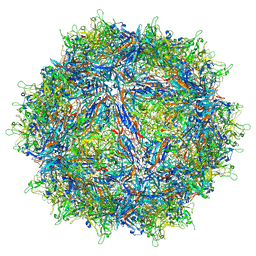 | | Structure of the adeno-associated virus 9 capsid at pH pH 7.4 in complex with terminal galactose | | Descriptor: | Capsid protein VP1, beta-D-galactopyranose | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-14 | | Release date: | 2021-06-09 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTG
 
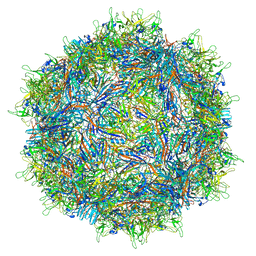 | | Structure of the adeno-associated virus 9 capsid at pH 6.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTP
 
 | | Structure of the adeno-associated virus 9 capsid at pH 5.5 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
7MTW
 
 | | Structure of the adeno-associated virus 9 capsid at pH 4.0 | | Descriptor: | Capsid protein VP1 | | Authors: | Penzes, J.J, Chipman, P, Bhattacharya, N, Zeher, A, Huang, R, McKenna, R, Agbandje-McKenna, M. | | Deposit date: | 2021-05-13 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Adeno-associated Virus 9 Structural Rearrangements Induced by Endosomal Trafficking pH and Glycan Attachment.
J.Virol., 95, 2021
|
|
1WM4
 
 | | Solution structure of mouse coactosin, an actin filament binding protein | | Descriptor: | Coactosin-like protein | | Authors: | Hellman, M, Paavilainen, V.O, Naumanen, P, Lappalainen, P, Annila, A, Permi, P. | | Deposit date: | 2004-07-03 | | Release date: | 2004-11-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of coactosin reveals structural homology to ADF/cofilin family proteins
Febs Lett., 576, 2004
|
|
