3IA3
 
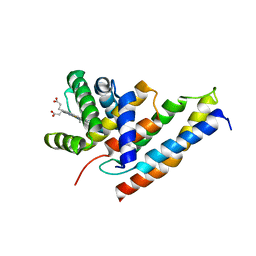 | |
2FMC
 
 | |
2QVT
 
 | | Structure of Melampsora lini avirulence protein, AvrL567-D | | Descriptor: | AvrL567-D | | Authors: | Guncar, G, Wang, C.I, Forwood, J.K, Teh, T, Catanzariti, A.M, Lawrence, G, Schirra, H.J, Anderson, P.A, Ellis, J.G, Dodds, P.N, Kobe, B. | | Deposit date: | 2007-08-08 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structures of flax rust avirulence proteins AvrL567-A and -D reveal details of the structural basis for flax disease resistance specificity.
Plant Cell, 19, 2007
|
|
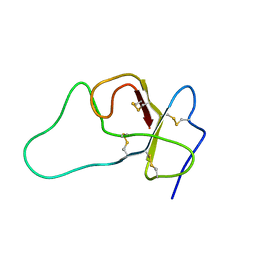
2QMS
 
 | | Crystal structure of a signaling molecule | | Descriptor: | Growth factor receptor-bound protein 7, SULFATE ION | | Authors: | Porter, C.J, Wilce, M.C, Wilce, J.A. | | Deposit date: | 2007-07-16 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation.
Bmc Struct.Biol., 7, 2007
|
|
2RGT
 
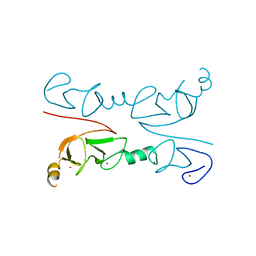 | | Crystal Structure of Lhx3 LIM domains 1 and 2 with the binding domain of Isl1 | | Descriptor: | Fusion of LIM/homeobox protein Lhx3, linker, Insulin gene enhancer protein ISL-1, ... | | Authors: | Bhati, M, Lee, M, Guss, J.M, Matthews, J.M. | | Deposit date: | 2007-10-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes.
Embo J., 27, 2008
|
|
8TOQ
 
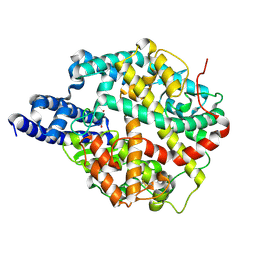 | | ACE2-peptide 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOS
 
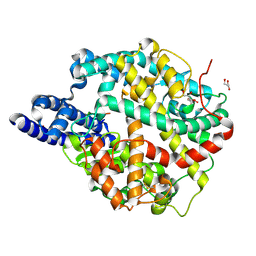 | | ACE2-peptide 6 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Christie, M, Payne, R. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOR
 
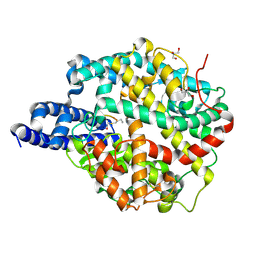 | | ACE2-peptide 2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOT
 
 | | ACE2-peptide2 complex crystal form 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Christie, M, Payne, R.J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
8TOU
 
 | | ACE2-peptide 2 complex crystal form 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Franck, C, Payne, R.J, Christie, M. | | Deposit date: | 2023-08-04 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of High Affinity Cyclic Peptide Ligands for Human ACE2 with SARS-CoV-2 Entry Inhibitory Activity.
Acs Chem.Biol., 19, 2024
|
|
2ZCG
 
 | |
1VTX
 
 | |
1EV0
 
 | |
1GL5
 
 | |
1HP3
 
 | |
1G9P
 
 | |
2K6A
 
 | | Solution structure of EAS D15 truncation mutant | | Descriptor: | Hydrophobin | | Authors: | Kwan, A.H. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity.
J.Mol.Biol., 382, 2008
|
|
2L4Z
 
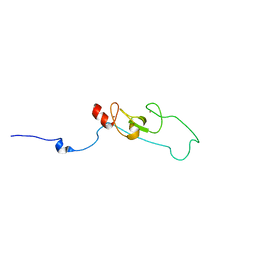 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
2OJP
 
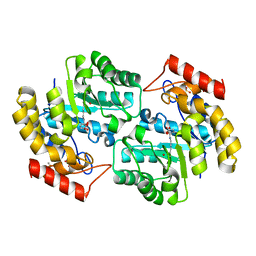 | | The crystal structure of a dimeric mutant of Dihydrodipicolinate synthase from E.coli- DHDPS-L197Y | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL | | Authors: | Griffin, M.D.W, Dobson, R.C.J, Antonio, L, Perugini, M.A, Jameson, G.B, Gerrard, J.A. | | Deposit date: | 2007-01-13 | | Release date: | 2008-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of quaternary structure in a homotetrameric enzyme.
J.Mol.Biol., 380, 2008
|
|
1RUT
 
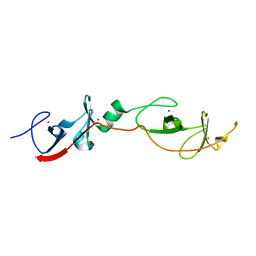 | | Complex of LMO4 LIM domains 1 and 2 with the ldb1 LID domain | | Descriptor: | Fusion protein of Lmo4 protein and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Ryan, D.P, Maher, M.J, Kwan, A.H.Y, Bacca, M, Mackay, J.P, Guss, J.M, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-12-11 | | Release date: | 2004-10-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Tandem LIM domains provide synergistic binding in the LMO4:Ldb1 complex
Embo J., 23, 2004
|
|
1JUN
 
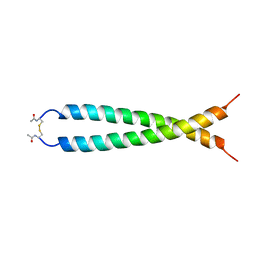 | |
2KWA
 
 | | 1H, 13C and 15N backbone and side chain resonance assignments of the N-terminal domain of the histidine kinase inhibitor KipI from Bacillus subtilis | | Descriptor: | Kinase A inhibitor | | Authors: | Hynson, R.M.G, Kwan, A, Jacques, D.A, Mackay, J.P, Trewhella, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Novel Structure of an Antikinase and its Inhibitor
J.Mol.Biol., 2010
|
|
2LSH
 
 | |
