1EKR
 
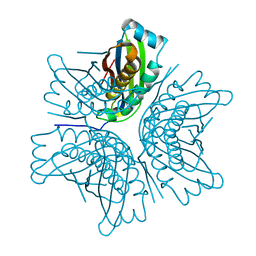 | | MOAC PROTEIN FROM E. COLI | | Descriptor: | MOLYBDENUM COFACTOR BIOSYNTHESIS PROTEIN C | | Authors: | Schindelin, H, Liu, M.T.W, Wuebbens, M.M, Rajagopalan, K.V. | | Deposit date: | 2000-03-09 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into molybdenum cofactor deficiency provided by the crystal structure of the molybdenum cofactor biosynthesis protein MoaC.
Structure Fold.Des., 8, 2000
|
|
1ORL
 
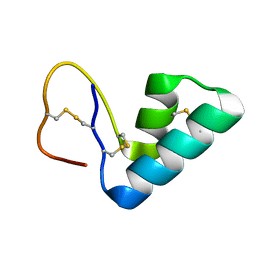 | | 1H NMR structure determination of Viscotoxin C1 | | Descriptor: | Viscotoxin C1 | | Authors: | Molinari, H, Romagnoli, S, Fogolari, F, Catalano, M, Urech, K, Giannattasio, M, Ragona, L. | | Deposit date: | 2003-03-14 | | Release date: | 2003-04-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of viscotoxin C1 from Viscum album species Coloratum ohwi: toward a structure-function analysis of viscotoxins.
Biochemistry, 42, 2003
|
|
8VER
 
 | |
2IMW
 
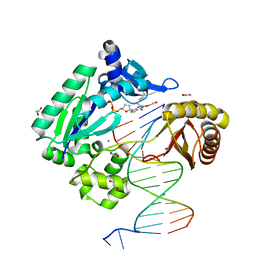 | | Mechanism of Template-Independent Nucleotide Incorporation Catalyzed by a Template-Dependent DNA Polymerase | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-dideoxyadenosine triphosphate, 5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3', ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2006-10-05 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of Template-independent Nucleotide Incorporation Catalyzed by a Template-dependent DNA Polymerase.
J.Mol.Biol., 365, 2007
|
|
3FDS
 
 | |
2AGQ
 
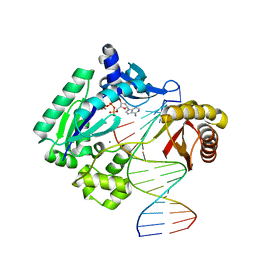 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*GP*GP*CP*TP*AP*CP*AP*GP*GP*AP*CP*TP*(DOC))-3', 5'-D(*TP*CP*AP*TP*GP*AP*GP*TP*CP*CP*TP*GP*TP*AP*GP*CP*C)-3', ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
2AGO
 
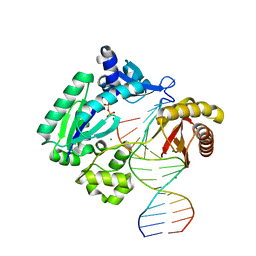 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*G)-3'), DNA (5'-D(*TP*TP*TP*TP*GP*AP*AP*TP*CP*CP*TP*TP*CP*CP*CP*CP*C)-3'), ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
2AGP
 
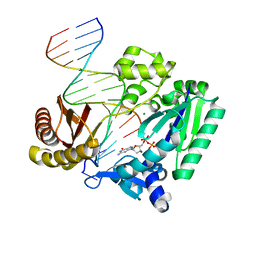 | | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*(DOC)-3'), ... | | Authors: | Ling, H, Yang, W. | | Deposit date: | 2005-07-27 | | Release date: | 2005-09-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis.
Embo J., 24, 2005
|
|
2B5E
 
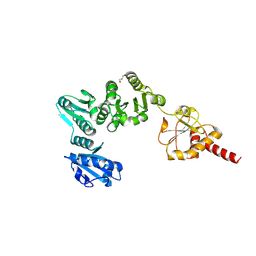 | | Crystal Structure of Yeast Protein Disulfide Isomerase | | Descriptor: | BARIUM ION, GLYCEROL, Protein disulfide-isomerase | | Authors: | Schindelin, H, Tian, G. | | Deposit date: | 2005-09-28 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of yeast protein disulfide isomerase suggests cooperativity between its active sites.
Cell(Cambridge,Mass.), 124, 2006
|
|
8YBQ
 
 | | Choline transporter BetT - CHT bound | | Descriptor: | BCCT family transporter, CHOLINE ION | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
8YBR
 
 | | Choline transporter BetT | | Descriptor: | BCCT family transporter | | Authors: | Yang, T.J, Nian, Y.W, Lin, H.J, Li, J, Zhang, J.R, Fan, M.R. | | Deposit date: | 2024-02-16 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structure and mechanism of the osmoregulated choline transporter BetT.
Sci Adv, 10, 2024
|
|
9CBT
 
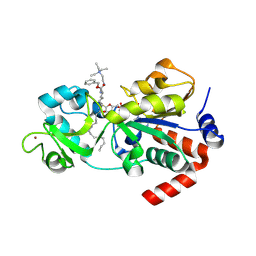 | | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10 | | Descriptor: | (phenylmethyl) ~{N}-[(2~{S})-6-[[(2~{R},3~{a}~{R},5~{R},6~{R},6~{a}~{R})-5-[[[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-6-oxidanyl-2-tridecyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxathiol-2-yl]amino]-1-oxidanylidene-1-[2-(triethyl-$l^{4}-azanyl)ethylamino]hexan-2-yl]carbamate, 2-{[(2S)-6-[(Z)-(1-{[(2R,3R,4R,5R)-5-({[(R)-{[(R)-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-4-hydroxy-2-sulfanyloxolan-3-yl]oxy}tetradecylidene)amino]-2-{[(benzyloxy)carbonyl]amino}hexanoyl]amino}-N,N,N-triethylethan-1-aminium (non-preferred name), NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Fenwick, M.K, Young, H.J, Lin, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human sirtuin 3 fragment (residues 118-399) bound to intermediates from reaction with NAD and inhibitor NH6-10
To Be Published
|
|
2DJY
 
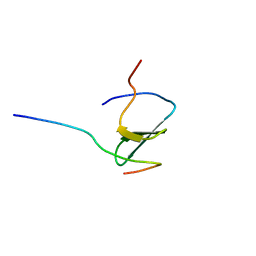 | | Solution structure of Smurf2 WW3 domain-Smad7 PY peptide complex | | Descriptor: | Mothers against decapentaplegic homolog 7, Smad ubiquitination regulatory factor 2 | | Authors: | Chong, P.A, Lin, H, Wrana, J.L, Forman-Kay, J.D. | | Deposit date: | 2006-04-06 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An Expanded WW Domain Recognition Motif Revealed by the Interaction between Smad7 and the E3 Ubiquitin Ligase Smurf2.
J.Biol.Chem., 281, 2006
|
|
6JZ6
 
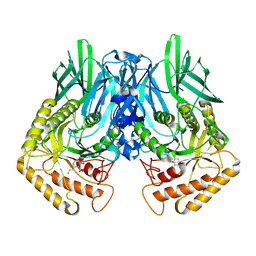 | | b-glucuronidase from Ruminococcus gnavus in complex with C6-substituted uronic isofagomine | | Descriptor: | (2~{S},3~{S},4~{R},5~{R})-4,5-bis(oxidanyl)-2-propyl-piperidine-3-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
4XBL
 
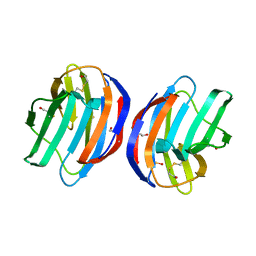 | |
4R8M
 
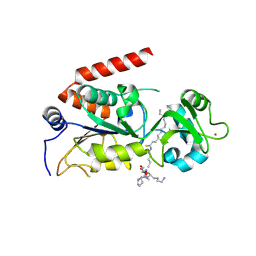 | | Human SIRT2 crystal structure in complex with BHJH-TM1 | | Descriptor: | BHJH-TM1 peptide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Teng, Y.B, Hao, Q, Lin, H.N, Jing, H. | | Deposit date: | 2014-09-02 | | Release date: | 2015-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Efficient Demyristoylase Activity of SIRT2 Revealed by Kinetic and Structural Studies
Sci Rep, 5, 2015
|
|
1CFF
 
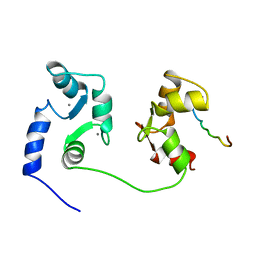 | | NMR SOLUTION STRUCTURE OF A COMPLEX OF CALMODULIN WITH A BINDING PEPTIDE OF THE CA2+-PUMP | | Descriptor: | CALCIUM ION, CALCIUM PUMP, CALMODULIN | | Authors: | Elshorst, B, Hennig, M, Foersterling, H, Diener, A, Maurer, M, Schulte, P, Schwalbe, H, Krebs, J, Schmid, H, Vorherr, T, Carafoli, E, Griesinger, C. | | Deposit date: | 1999-03-18 | | Release date: | 1999-09-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump.
Biochemistry, 38, 1999
|
|
6JJC
 
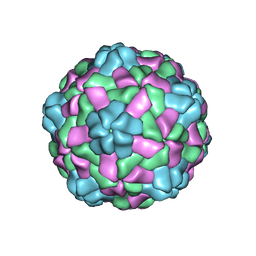 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) semi-empty VLP
To Be Published
|
|
6JJD
 
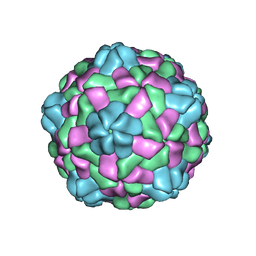 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii nodavirus (MrNV) full VLP
To Be Published
|
|
6JJA
 
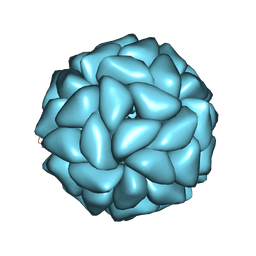 | | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP | | Descriptor: | CALCIUM ION, Nucleocapsid protein CP17 | | Authors: | Chang, W.H, Wang, C.H, Lin, H.H, Lin, S.Y, Chong, S.C, Wu, Y.Y. | | Deposit date: | 2019-02-25 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Cryo-EM structure of giant freshwater prawn Macrobrachium rosenbergii extra small virus (XSV) VLP
To Be Published
|
|
6JZ2
 
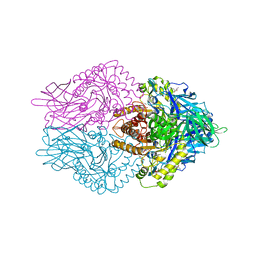 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic isofagomine at 1.3 Angstroms resolution | | Descriptor: | (3S,4R,5R)-4,5-dihydroxypiperidine-3-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ8
 
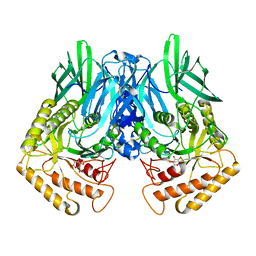 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro 1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.583 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ5
 
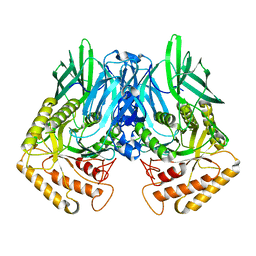 | |
6JZ3
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with uronic deoxynojirimycin | | Descriptor: | (2~{S},3~{R},4~{R},5~{S})-3,4,5-tris(oxidanyl)piperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
6JZ4
 
 | | b-glucuronidase from Ruminococcus gnavus in complex with D-glucaro-d-lactam | | Descriptor: | (2S,3R,4S,5R)-3,4,5-trihydroxy-6-oxopiperidine-2-carboxylic acid, (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-glucuronidase | | Authors: | Dashnyam, P, Lin, H.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Substituent Position of Iminocyclitols Determines the Potency and Selectivity for Gut Microbial Xenobiotic-Reactivating Enzymes.
J.Med.Chem., 63, 2020
|
|
