5VKC
 
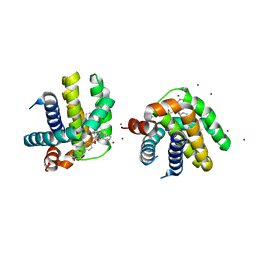 | | Crystal structure of MCL-1 in complex with a BIM competitive inhibitor | | Descriptor: | 7-(3-{[4-(4-acetylpiperazin-1-yl)phenoxy]methyl}-1,5-dimethyl-1H-pyrazol-4-yl)-3-{3-[(naphthalen-1-yl)oxy]propyl}-1-[(pyridin-3-yl)methyl]-1H-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Judge, R.A, Souers, A.J. | | Deposit date: | 2017-04-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure-guided design of a series of MCL-1 inhibitors with high affinity and selectivity.
J. Med. Chem., 58, 2015
|
|
8WMH
 
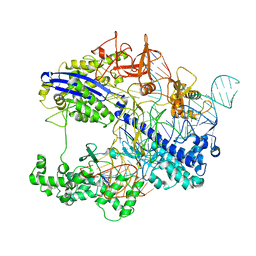 | | Structure of CbCas9 bound to 6-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-03 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMM
 
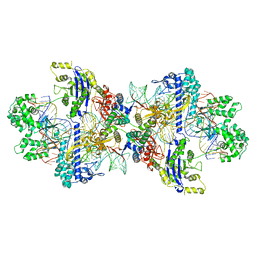 | | Structure of CbCas9-PcrIIC1 complex bound to 28-bp DNA substrate (20-nt complementary) | | Descriptor: | MAGNESIUM ION, NTS, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WMN
 
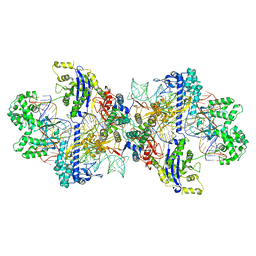 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (symmetric 20-nt complementary) | | Descriptor: | DNA (62-MER), MAGNESIUM ION, PcrIIC1, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8WR4
 
 | | Structure of CbCas9-PcrIIC1 complex bound to 62-bp DNA substrate (non-targeting complex) | | Descriptor: | CbCas9 effector-1, DNA (62-MER), MAGNESIUM ION, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-10-13 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
5IAW
 
 | | Novel natural FXR modulator with a unique binding mode | | Descriptor: | (1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl 4-hydroxybenzoate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
5ICK
 
 | | A unique binding model of FXR LBD with feroline | | Descriptor: | (1S,2S,3Z,5S,8Z)-5-hydroxy-5,9-dimethyl-2-(propan-2-yl)cyclodeca-3,8-dien-1-yl 4-hydroxybenzoate, Bile acid receptor, Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-23 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
8EFK
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with hairpin DNA | | Descriptor: | 2',3'-dideoxyadenosine triphosphate, DNA (5'-D(P*TP*TP*TP*TP*GP*GP*CP*TP*TP*TP*TP*GP*CP*CP*(2DA))-3'), Lates calcarifer DNA polymerase theta, ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8EF9
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*C)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8EFC
 
 | | Structure of Lates calcarifer DNA polymerase theta polymerase domain with long duplex DNA, complex Ia | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*CP*TP*GP*TP*GP*AP*GP*GP*CP*AP*TP*CP*CP*GP*TP*AP*GP*(2DA))-3'), DNA (5'-D(*AP*GP*CP*TP*CP*TP*AP*CP*GP*GP*AP*TP*GP*CP*CP*TP*CP*AP*CP*AP*G)-3'), ... | | Authors: | Li, C, Zhu, H, Sun, J, Gao, Y. | | Deposit date: | 2022-09-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of DNA polymerase theta mediated DNA end joining.
Nucleic Acids Res., 51, 2023
|
|
8GHX
 
 | | Crystal Structure of CelD Cellulase from the Anaerobic Fungus Piromyces finnis | | Descriptor: | 1,2-ETHANEDIOL, Cellulase CelD | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
8GHY
 
 | | Crystal Structure of the E154D mutant CelD Cellulase from the Anaerobic Fungus Piromyces finnis in the complex with cellotriose. | | Descriptor: | Cellulase CelD, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Dementieve, A, Kim, Y, Jedrzejczak, R, Michalska, K, Joachimiak, A. | | Deposit date: | 2023-03-13 | | Release date: | 2023-05-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and enzymatic characterization of CelD endoglucanase from the anaerobic fungus Piromyces finnis.
Appl.Microbiol.Biotechnol., 107, 2023
|
|
1F8Q
 
 | | CRYSTAL STRUCTURE OF ALPHA-MOMORCHARIN IN ACETONITRILE-WATER MIXTURE | | Descriptor: | ACETONITRILE, ALPHA-MOMORCHARIN, PENTANEDIAL, ... | | Authors: | Zhu, G, Huang, Q, Qian, M, Tang, Y. | | Deposit date: | 2000-07-03 | | Release date: | 2000-07-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of alpha-momorcharin in 80% acetonitrile--water mixture
BIOCHIM.BIOPHYS.ACTA, 1548, 2001
|
|
3U2Z
 
 | | Activator-Bound Structure of Human Pyruvate Kinase M2 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-(3-aminobenzyl)-4-methyl-2-methylsulfinyl-4,6-dihydro-5H-thieno[2',3':4,5]pyrrolo[2,3-d]pyridazin-5-one, Pyruvate kinase isozymes M1/M2, ... | | Authors: | Hong, B, Dimov, S, Tempel, W, Auld, D, Thomas, C, Boxer, M, Jianq, J.-K, Skoumbourdis, A, Min, S, Southall, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Inglese, J, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-10-04 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis.
Nat.Chem.Biol., 8, 2012
|
|
4HVI
 
 | |
4HVG
 
 | |
6M6B
 
 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase and ATP-gamma-S | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
4HVH
 
 | |
7LH7
 
 | | Crystal structure of BCL-XL in complex with a benzothiazole-based inhibitor | | Descriptor: | Bcl-2-like protein 1, N-(1,3-benzothiazol-2-yl)-2-(4-{[(4-{[(2R)-4-(morpholin-4-yl)-1-(phenylsulfanyl)butan-2-yl]amino}-3-[(trifluoromethyl)sulfonyl]phenyl)sulfonyl]carbamoyl}-1,3-thiazol-2-yl)-1,2,3,4-tetrahydroisoquinoline-8-carboxamide | | Authors: | Judge, R.A, Tao, Z. | | Deposit date: | 2021-01-21 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.409 Å) | | Cite: | Structure-Based Design of A-1293102, a Potent and Selective BCL-XL Inhibitor
ACS Medicinal Chemistry Letters, 12, 2021
|
|
4HVD
 
 | | JAK3 kinase domain in complex with 2-Cyclopropyl-5H-pyrrolo[2,3-b]pyrazine-7-carboxylic acid ((S)-1,2,2-trimethyl-propyl)-amide | | Descriptor: | 1-phenylurea, 2-cyclopropyl-N-[(2S)-3,3-dimethylbutan-2-yl]-5H-pyrrolo[2,3-b]pyrazine-7-carboxamide, Tyrosine-protein kinase JAK3 | | Authors: | Kuglstatter, A, Shao, A. | | Deposit date: | 2012-11-06 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 3-Amido Pyrrolopyrazine JAK Kinase Inhibitors: Development of a JAK3 vs JAK1 Selective Inhibitor and Evaluation in Cellular and in Vivo Models.
J.Med.Chem., 56, 2013
|
|
6M6C
 
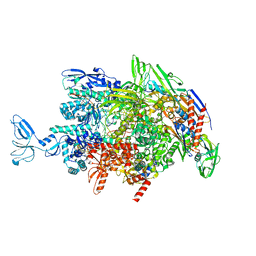 | | CryoEM structure of Thermus thermophilus RNA polymerase elongation complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
3INM
 
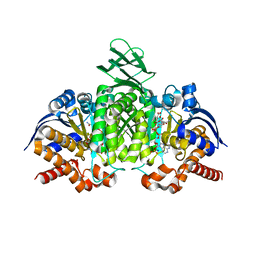 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase R132H mutant in complex with NADPH, ALPHA-KETOGLUTARATE and CALCIUM(2+) | | Descriptor: | 2-OXOGLUTARIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Fontano, E, Brown, R.S, Suto, R.K, Bhyravbhatla, B. | | Deposit date: | 2009-08-12 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature, 462, 2009
|
|
6M6A
 
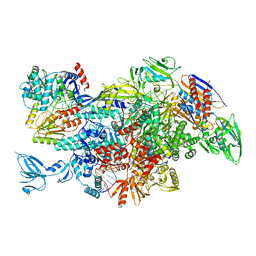 | | Cryo-EM structure of Thermus thermophilus Mfd in complex with RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Shi, J, Wen, A, Feng, Y. | | Deposit date: | 2020-03-14 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural basis of Mfd-dependent transcription termination.
Nucleic Acids Res., 48, 2020
|
|
9JDZ
 
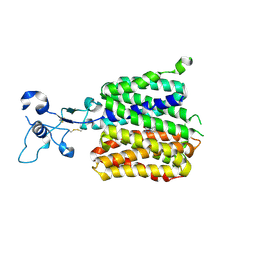 | | Human URAT1 bound to lesinurad | | Descriptor: | Solute carrier family 22 member 12, lesinurad | | Authors: | Wu, C, Xu, H.E. | | Deposit date: | 2024-09-01 | | Release date: | 2024-10-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Molecular mechanisms of uric acid transport by the native human URAT1 and its inhibition by anti-gout drugs
Biorxiv, 2024
|
|
7YTO
 
 | |
