4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4IPA
 
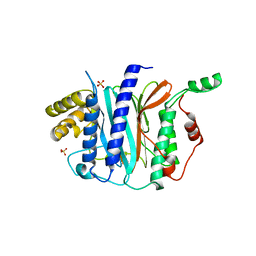 | | Structure of a thermophilic Arx1 | | Descriptor: | Putative curved DNA-binding protein, SULFATE ION | | Authors: | Bange, G, Sinning, I. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Consistent mutational paths predict eukaryotic thermostability.
BMC Evol Biol, 13, 2013
|
|
3ZRI
 
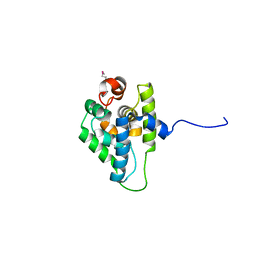 | |
3ZRJ
 
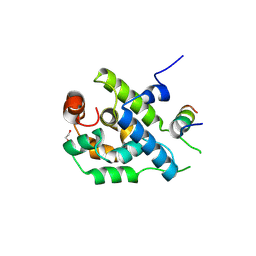 | | Complex of ClpV N-domain with VipB peptide | | Descriptor: | 1,2-ETHANEDIOL, CLPB PROTEIN, VIPB | | Authors: | Lenherr, E.D, Kopp, J, Sinning, I. | | Deposit date: | 2011-06-16 | | Release date: | 2011-07-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Molecular Basis for the Unique Role of the Aaa+ Chaperone Clpv in Type Vi Protein Secretion.
J.Biol.Chem., 286, 2011
|
|
7AQE
 
 | | Structure of SARS-CoV-2 Main Protease bound to UNC-2327 | | Descriptor: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P, Meents, A. | | Deposit date: | 2020-10-21 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6Y0H
 
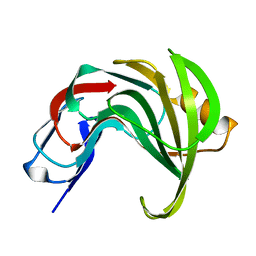 | | High resolution structure of GH11 xylanase from Nectria haematococca | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Andaleeb, H, Betzel, C, Perbandt, M, Brognaro, H. | | Deposit date: | 2020-02-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structure and biochemical characterization of a GH11 endoxylanase from Nectria haematococca.
Sci Rep, 10, 2020
|
|
9EUN
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with SAM and m7GTP | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMV
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-11 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EMJ
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with Toyocamycin and m7GpppA (Cap0-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
9EML
 
 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and m7GpppA (Cap0-analog)/m7GpppAm (Cap1-analog) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Kiene, A. | | Deposit date: | 2024-03-08 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
7ALZ
 
 | | GqqA- a novel type of quorum quenching acylases | | Descriptor: | PHENYLALANINE, Prephenate dehydratase | | Authors: | Werner, N, Betzel, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity.
Sci Rep, 11, 2021
|
|
7AM0
 
 | | GqqA- a novel type of quorum quenching acylases | | Descriptor: | PHENYLALANINE, Prephenate dehydratase | | Authors: | Werner, N, Betzel, C. | | Deposit date: | 2020-10-07 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Komagataeibacter europaeus GqqA is the prototype of a novel bifunctional N-Acyl-homoserine lactone acylase with prephenate dehydratase activity.
Sci Rep, 11, 2021
|
|
8S8W
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Sangivamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
8S8X
 
 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with Toyocamycin and m7GpppA-RNA (Cap0-RNA) | | Descriptor: | 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D, Scheer, T.E.S. | | Deposit date: | 2024-03-07 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug development
To Be Published
|
|
8CJ4
 
 | | Crystal structure of ClpP from Staphylococcus epidermidis, tetradecamer | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit | | Authors: | Alves Franca, B, Rohde, H, Betzel, C. | | Deposit date: | 2023-02-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular insights into the dynamic modulation of bacterial ClpP function and oligomerization by peptidomimetic boronate compounds.
Sci Rep, 14, 2024
|
|
7PXZ
 
 | | Reduced form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-08 | | Release date: | 2023-01-18 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
7PZQ
 
 | | Oxidized form of SARS-CoV-2 Main Protease determined by XFEL radiation | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Schubert, R, Reinke, P, Galchenkova, M, Oberthuer, D, Murillo, G.E.P, Kim, C, Bean, R, Turk, D, Hinrichs, W, Middendorf, P, Round, A, Schmidt, C, Mills, G, Kirkwood, H, Han, H, Koliyadu, J, Bielecki, J, Gelisio, L, Sikorski, M, Kloos, M, Vakilii, M, Yefanov, O.N, Vagovic, P, de-Wijn, R, Letrun, R, Guenther, S, White, T.A, Sato, T, Srinivasan, V, Kim, Y, Chretien, A, Han, S, Brognaro, H, Maracke, J, Knoska, J, Seychell, B.C, Brings, L, Norton-Baker, B, Geng, T, Dore, A.S, Uetrecht, C, Redecke, L, Beck, T, Lorenzen, K, Betzel, C, Mancuso, A.P, Bajt, S, Chapman, H.N, Meents, A, Lane, T.J. | | Deposit date: | 2021-10-13 | | Release date: | 2023-01-25 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | SARS-CoV-2 M pro responds to oxidation by forming disulfide and NOS/SONOS bonds.
Nat Commun, 15, 2024
|
|
6FDG
 
 | |
8BS0
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 80 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRZ
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at 52 MPa helium gas pressure in a sapphire capillary | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BRY
 
 | | Room-temperature structure of Pedobacter heparinus N-acetylglucosamine 2-epimerase at atmospheric pressure | | Descriptor: | CHLORIDE ION, N-acylglucosamine 2-epimerase, PHOSPHATE ION | | Authors: | Lieske, J, Saouane, S, Assmann, M, Zaun, H, Kuballa, J, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BS1
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at atmospheric pressure | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8BS2
 
 | | Room-temperature structure of SARS-CoV-2 Main protease at 104 MPa helium gas pressure in a sapphire capillary | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lieske, J, Saouane, S, Guenther, S, Reinke, P.Y.A, Rahmani Mashhour, A, Meents, A. | | Deposit date: | 2022-11-24 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | High-pressure macromolecular crystallography to explore the conformational space of proteins
To Be Published
|
|
8QYF
 
 | |
8JEE
 
 | | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, Levosulpiride, ... | | Authors: | Rasheed, S, Huda, N, Falke, S, Fisher, S.Z, Ahmad, M.S. | | Deposit date: | 2023-05-15 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Crystal Structure of Human Carbonic Anhydrase II In-complex with Levosulpiride at 2.96 A Resolution
To Be Published
|
|
