6DE8
 
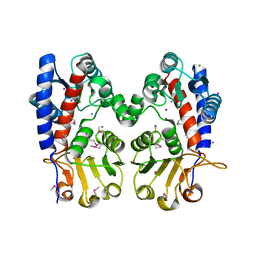 | | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase from Campylobacter jejuni | | Descriptor: | Bifunctional protein FolD, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Zhang, R, Peterson, S.N, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-05-11 | | Release date: | 2018-05-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Crystal Structure of Bifunctional Enzyme FolD-Methylenetetrahydrofolate Dehydrogenase/Cyclohydrolase from Campylobacter jejuni
To Be Published
|
|
6X4I
 
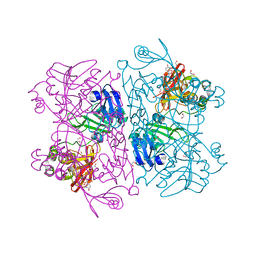 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with 3'-uridinemonophosphate | | Descriptor: | 1,2-ETHANEDIOL, 3'-URIDINEMONOPHOSPHATE, SODIUM ION, ... | | Authors: | Chang, C, Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
8CRV
 
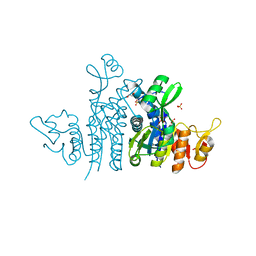 | | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, Carbamate kinase, FORMIC ACID, ... | | Authors: | Kim, Y, Skarina, T, Mesa, N, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Carbamate Kinase from Pseudomonas aeruginosa
To Be Published
|
|
8EBC
 
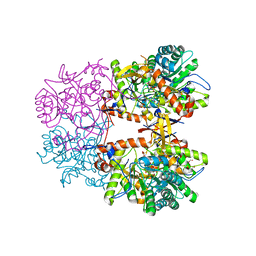 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP | | Descriptor: | FORMIC ACID, GLYCEROL, INOSINIC ACID, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Osipiuk, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-08-31 | | Release date: | 2022-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria monocytogenes in the complex with IMP
To Be Published
|
|
7RLR
 
 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RL8
 
 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | Descriptor: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2021-07-23 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7R6S
 
 | | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia | | Descriptor: | Putative bacteriophage protein, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Brunzelle, J.S, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2021-06-23 | | Release date: | 2022-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Putative Bacteriophage Protein from Stenotrophomonas maltophilia
To Be Published
|
|
3HFR
 
 | | Crystal structure of glutamate racemase from Listeria monocytogenes | | Descriptor: | CHLORIDE ION, Glutamate racemase, O-(O-(2-AMINOPROPYL)-O'-(2-METHOXYETHYL)POLYPROPYLENE GLYCOL 500) | | Authors: | Majorek, K.A, Chruszcz, M, Zimmerman, M.D, Klimecka, M.M, Cymborowski, M, Skarina, T, Onopriyenko, O, Stam, J, Otwinowski, Z, Anderson, W.F, Savchenko, A, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-12 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of glutamate racemase from Listeria monocytogenes
TO BE PUBLISHED
|
|
3HJV
 
 | | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, CHLORIDE ION, Malonyl Coa-acyl carrier protein transacylase, ... | | Authors: | Halavaty, A.S, Wawrzak, Z, Anderson, S, Skarina, T, Onopriyenko, O, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom resolution crystal structure of an acyl carrier protein S-malonyltransferase from Vibrio cholerae O1 biovar eltor str. N16961
To be Published
|
|
3HJB
 
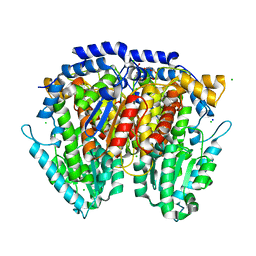 | | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-21 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Glucose-6-phosphate Isomerase from Vibrio cholerae.
To be Published
|
|
3IJ3
 
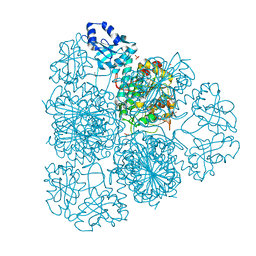 | | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cytosol aminopeptidase, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Cytosol Aminopeptidase from Coxiella burnetii
TO BE PUBLISHED
|
|
3IMF
 
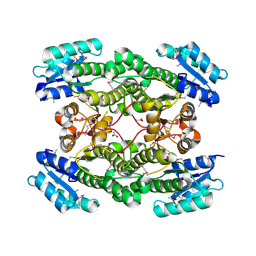 | | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | ACETATE ION, Short chain dehydrogenase | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Peterson, S, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 1.99 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3IAC
 
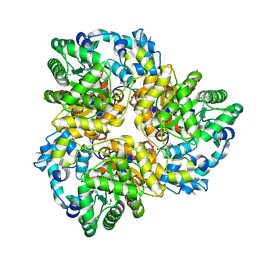 | | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium. | | Descriptor: | CHLORIDE ION, Glucuronate isomerase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-13 | | Release date: | 2009-07-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 2.2 Angstrom Crystal Structure of Glucuronate Isomerase from Salmonella typhimurium.
To be Published
|
|
3IFE
 
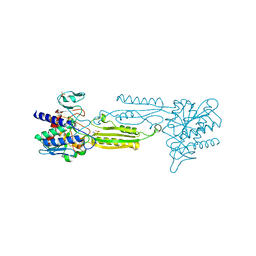 | | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'. | | Descriptor: | Peptidase T, SODIUM ION, SULFATE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | 1.55 Angstrom Resolution Crystal Structure of Peptidase T (pepT-1) from Bacillus anthracis str. 'Ames Ancestor'.
To be Published
|
|
3IHS
 
 | | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Brunzelle, J.S, Anderson, S.M, Skarina, T, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of a Phosphocarrier Protein HPr from Bacillus anthracis str. Ames
To be Published
|
|
3IB3
 
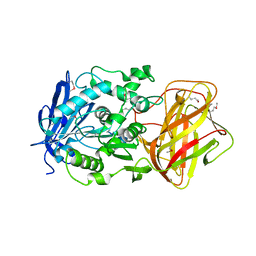 | | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Domagalski, M.J, Chruszcz, M, Osinski, T, Skarina, T, Onopriyenko, O, Cymborowski, M, Shumilin, I.A, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-15 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of SACOL2612 - CocE/NonD family hydrolase from Staphylococcus aureus
To be Published
|
|
3III
 
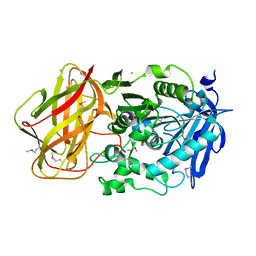 | | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus | | Descriptor: | CHLORIDE ION, CocE/NonD family hydrolase, NICKEL (II) ION, ... | | Authors: | Osinski, T, Chruszcz, M, Domagalski, M.J, Cymborowski, M, Shumilin, I.A, Skarina, T, Onopriyenko, O, Zimmerman, M.D, Savchenko, A, Edwards, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-01 | | Release date: | 2009-08-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Crystal Structure of CocE/NonD family hydrolase (SACOL2612) from Staphylococcus aureus
To be Published
|
|
3IFS
 
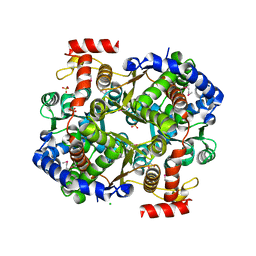 | | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Glucose-6-phosphate isomerase, LITHIUM ION, ... | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-24 | | Release date: | 2009-08-11 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | 2.0 Angstrom Resolution Crystal Structure of Glucose-6-phosphate Isomerase (pgi) from Bacillus anthracis.
To be Published
|
|
3IR4
 
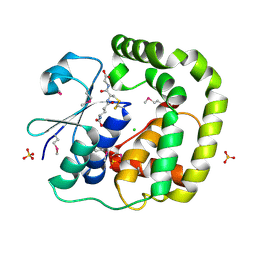 | | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione | | Descriptor: | CHLORIDE ION, GLUTATHIONE, Glutaredoxin 2, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Halavaty, A, Dauter, Z, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-01 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | 1.2 Angstrom Crystal Structure of the Glutaredoxin 2 (grxB) from Salmonella typhimurium in complex with Glutathione.
TO BE PUBLISHED
|
|
3IG4
 
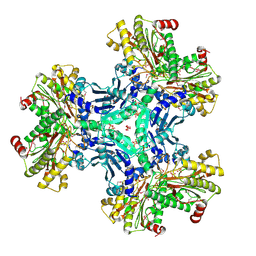 | | Structure of a putative aminopeptidase P from Bacillus anthracis | | Descriptor: | MANGANESE (II) ION, SULFATE ION, Xaa-pro aminopeptidase | | Authors: | Anderson, S.M, Wawrzak, Z, Skarina, T, Onopriyenko, O, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a putative aminopeptidase P from Bacillus anthracis
To be Published
|
|
3IMI
 
 | | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor' | | Descriptor: | HIT family protein, SULFATE ION, ZINC ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Gordon, E, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-10 | | Release date: | 2009-08-18 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2.01 Angstrom resolution crystal structure of a HIT family protein from Bacillus anthracis str. 'Ames Ancestor'
To be Published
|
|
3IJ5
 
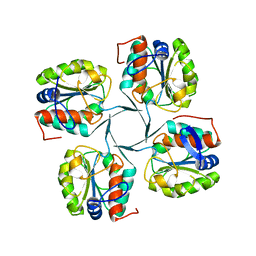 | | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis | | Descriptor: | 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase, CHLORIDE ION | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-03 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase from Yersinia pestis
TO BE PUBLISHED
|
|
3IJR
 
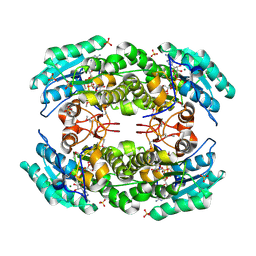 | | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+ | | Descriptor: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase, ... | | Authors: | Halavaty, A.S, Minasov, G, Skarina, T, Onopriyenko, O, Gordon, E, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom resolution crystal structure of a short chain dehydrogenase from Bacillus anthracis str. 'Ames Ancestor' in complex with NAD+
To be Published
|
|
3INP
 
 | | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis. | | Descriptor: | CHLORIDE ION, D-ribulose-phosphate 3-epimerase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Scott, P, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of D-ribulose-phosphate 3-epimerase from Francisella tularensis.
TO BE PUBLISHED
|
|
3IGX
 
 | | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis. | | Descriptor: | PHOSPHATE ION, Transaldolase | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Gordon, E, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-29 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 1.85 Angstrom Resolution Crystal Structure of Transaldolase B (talA) from Francisella tularensis.
TO BE PUBLISHED
|
|
