3VRI
 
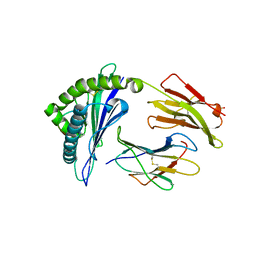 | | HLA-B*57:01-RVAQLENVYI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
6KJZ
 
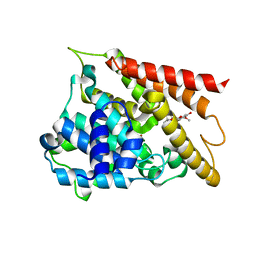 | | Crystal structure of PDE4D catalytic domain complexed with compound 1 | | Descriptor: | 5,9-dihydroxy-8-methoxy-2,2-dimethyl-7-(3-methylbut-2-en-1-yl)-3,4-dihydro-2H,6H-pyrano[3,2-b]xanthen-6-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y.-Y, He, X, Luo, H.-B. | | Deposit date: | 2019-07-23 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.200001 Å) | | Cite: | Discovery and Optimization of alpha-Mangostin Derivatives as Novel PDE4 Inhibitors for the Treatment of Vascular Dementia.
J.Med.Chem., 63, 2020
|
|
6N05
 
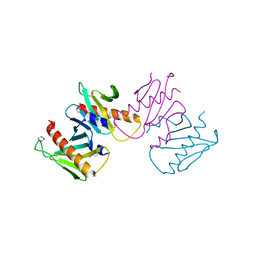 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
4Y3U
 
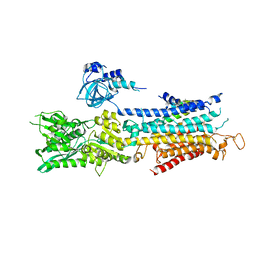 | | The structure of phospholamban bound to the calcium pump SERCA1a | | Descriptor: | Cardiac phospholamban, POTASSIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Hurley, T.D. | | Deposit date: | 2015-02-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
J. Biol. Chem., 288, 2013
|
|
3VFU
 
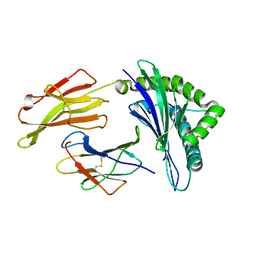 | | crystal structure of HLA B*3508 LPEP-P7Ala, peptide mutant P7-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P7A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
6LHD
 
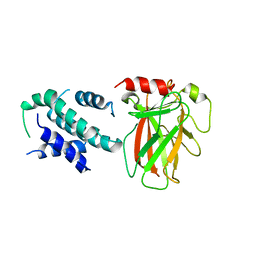 | | Crystal structure of p53/BCL-xL fusion complex | | Descriptor: | ZINC ION, fusion protein of Bcl-2-like protein 1 and Isoform 6 of Cellular tumor antigen p53 | | Authors: | Wei, H, Chen, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural insight into the molecular mechanism of p53-mediated mitochondrial apoptosis.
Nat Commun, 12, 2021
|
|
3VFS
 
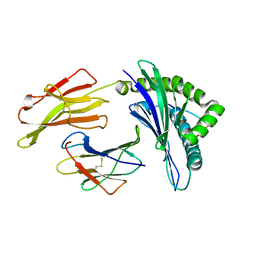 | | crystal structure of HLA B*3508LPEP-P5Ala , peptide mutant P5-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P5A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
6KW4
 
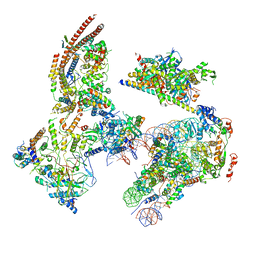 | | The ClassB RSC-Nucleosome Complex | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC3, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-09-06 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (7.55 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
4KYT
 
 | | The structure of superinhibitory phospholamban bound to the calcium pump SERCA1a | | Descriptor: | Cardiac phospholamban, POTASSIUM ION, SERCA1a, ... | | Authors: | Hurley, T.D, Akin, B.L, Jones, L.R. | | Deposit date: | 2013-05-29 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
J.Biol.Chem., 288, 2013
|
|
3VFP
 
 | | crystal structure of HLA B*3508 LPEP158G, HLA mutant Gly158 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, LPEP peptide from EBV, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
7C84
 
 | | Esterase AlinE4 mutant, D162A | | Descriptor: | ACETATE ION, CADMIUM ION, GLYCEROL, ... | | Authors: | Li, Z, Li, J. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Structure-guided protein engineering increases enzymatic activities of the SGNH family esterases.
Biotechnol Biofuels, 13, 2020
|
|
5W0Q
 
 | | CREBBP Bromodomain in complex with Cpd17 (N,2,7-trimethyl-2,3-dihydro-4H-benzo[b][1,4]oxazine-4-carboxamide) | | Descriptor: | (2R)-N,2,7-trimethyl-2,3-dihydro-4H-1,4-benzoxazine-4-carboxamide, CREB-binding protein, SULFATE ION | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
6K15
 
 | | RSC substrate-recruitment module | | Descriptor: | Chromatin structure-remodeling complex protein RSC3, Chromatin structure-remodeling complex protein RSC30, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Ye, Y.P, Wu, H, Chen, K.J, Verma, N, Cairns, B, Gao, N, Chen, Z.C. | | Deposit date: | 2019-05-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the RSC complex bound to the nucleosome.
Science, 366, 2019
|
|
2OCV
 
 | | Structural basis of Na+ activation mimicry in murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
5W0F
 
 | | CREBBP Bromodomain in complex with Cpd3 ((S)-1-(3-(6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydrofuran-3-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-[(3S)-oxolan-3-yl]-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-30 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
5W0L
 
 | | CREBBP Bromodomain in complex with Cpd10 (1-(3-(7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl)-1-(tetrahydro-2H-pyran-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl)ethan-1-one) | | Descriptor: | 1-{3-[7-(difluoromethyl)-6-(1-methyl-1H-pyrazol-4-yl)-3,4-dihydroquinolin-1(2H)-yl]-1-(oxan-4-yl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl}ethan-1-one, CREB-binding protein | | Authors: | Murray, J.M. | | Deposit date: | 2017-05-31 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | GNE-781, A Highly Advanced Potent and Selective Bromodomain Inhibitor of Cyclic Adenosine Monophosphate Response Element Binding Protein, Binding Protein (CBP).
J. Med. Chem., 60, 2017
|
|
3QSM
 
 | | Crystal structure for the MSOX.chloride binary complex | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
3VFT
 
 | | crystal structure of HLA B*3508LPEP-P6Ala, peptide mutant P6-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P6A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4HY1
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-({4-[(3R)-3-aminopyrrolidin-1-yl]-5-chloro-6-ethyl-7H-pyrrolo[2,3-d]pyrimidin-2-yl}sulfanyl)-1H-isoindol-1-one, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3VRJ
 
 | | HLA-B*57:01-LTTKLTNTNI in complex with abacavir | | Descriptor: | 10-mer peptide, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Vivian, J.P, Illing, P.T, McCluskey, J, Rossjohn, J. | | Deposit date: | 2012-04-11 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immune self-reactivity triggered by drug-modified HLA-peptide repertoire
Nature, 486, 2012
|
|
3VFR
 
 | | crystal structure of HLA B*3508LPEP-P4Ala, peptide mutant P4-ala | | Descriptor: | Beta-2-microglobulin, LPEP peptide from EBV, P4A, ... | | Authors: | Liu, Y.C, Rossjohn, J, Gras, S. | | Deposit date: | 2012-01-10 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Energetic Basis Underpinning T-cell Receptor Recognition of a Super-bulged Peptide Bound to a Major Histocompatibility Complex Class I Molecule.
J.Biol.Chem., 287, 2012
|
|
4LCW
 
 | | The structure of human MAIT TCR in complex with MR1-K43A-RL-6-Me-7OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Rossjohn, J. | | Deposit date: | 2013-06-24 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antigen-loaded MR1 tetramers define T cell receptor heterogeneity in mucosal-associated invariant T cells.
J.Exp.Med., 210, 2013
|
|
3QSE
 
 | | Crystal structure for the complex of substrate-reduced msox with sarcosine | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Monomeric sarcosine oxidase, ... | | Authors: | Kommoju, P, Chen, Z, Bruckner, R.C, Mathews, F.S, Jorns, M.S. | | Deposit date: | 2011-02-21 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing oxygen activation sites in two flavoprotein oxidases using chloride as an oxygen surrogate.
Biochemistry, 50, 2011
|
|
4HXZ
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | 6-ethyl-4-methoxy-2-(pyridin-3-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbaldehyde, Topoisomerase IV, subunit B | | Authors: | Bensen, D.C, Creighton, C.J, Kwan, B, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HXW
 
 | | Pyrrolopyrimidine inhibitors of dna gyrase b and topoisomerase iv, part i: structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity. | | Descriptor: | (3R)-1-[5-chloro-6-ethyl-2-(pyrido[2,3-b]pyrazin-7-ylsulfanyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]pyrrolidin-3-amine, DNA gyrase subunit B, TERTIARY-BUTYL ALCOHOL | | Authors: | Bensen, D.C, Trzoss, M, Tari, L.W. | | Deposit date: | 2012-11-12 | | Release date: | 2013-02-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Pyrrolopyrimidine inhibitors of DNA gyrase B (GyrB) and topoisomerase IV (ParE). Part I: Structure guided discovery and optimization of dual targeting agents with potent, broad-spectrum enzymatic activity.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
