3RQQ
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis in complex with P1,P3-Di(adenosine-5') triphosphate | | Descriptor: | ADP/ATP-DEPENDENT NAD(P)H-HYDRATE DEHYDRATASE, BIS(ADENOSINE)-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RQ8
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with P1,P5-Di(adenosine-5') pentaphosphate | | Descriptor: | ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RXY
 
 | | Crystal structure of NIF3 superfamily protein from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Michalska, K, Tesar, C, Clancy, S, Otwinowski, Z, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-10 | | Release date: | 2011-06-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NIF3 superfamily protein from Sphaerobacter thermophilus
To be Published
|
|
3SBL
 
 | | Crystal Structure of New Delhi Metal-beta-lactamase-1 from Klebsiella pneumoniae | | Descriptor: | Beta-lactamase NDM-1, CITRIC ACID | | Authors: | Kim, Y, Tesar, C, Jedrzejczak, R, Babnigg, J, Binkowski, T.A, Mire, J, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2011-06-05 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structure of Apo- and Monometalated Forms of NDM-1 A Highly Potent Carbapenem-Hydrolyzing Metallo-beta-Lactamase
Plos One, 6, 2011
|
|
3TJY
 
 | | Structure of the Pto-binding domain of HopPmaL generated by limited chymotrypsin digestion | | Descriptor: | CHLORIDE ION, Effector protein hopAB3, SULFATE ION | | Authors: | Singer, A.U, Stein, A, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-08-25 | | Release date: | 2011-09-14 | | Last modified: | 2013-01-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of HopPmaL reveals the presence of a second adaptor domain common to the HopAB family of Pseudomonas syringae type III effectors.
Biochemistry, 51, 2012
|
|
3RNS
 
 | |
3RQZ
 
 | | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus | | Descriptor: | ACETATE ION, Metallophosphoesterase, ZINC ION | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of metallophosphoesterase from Sphaerobacter thermophilus
To be Published
|
|
3RHF
 
 | | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CITRATE ANION, ... | | Authors: | Nocek, B, Hatzos-Skintges, C, Feldmann, B, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-11 | | Release date: | 2011-06-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Polyphosphate Kinase 2 from Arthrobacter aurescens TC1
TO BE PUBLISHED
|
|
3RQ6
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis soaked with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, MAGNESIUM ION | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3RMQ
 
 | | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis (V71M mutant) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ZINC ION, ... | | Authors: | Michalska, K, Weger, A, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-21 | | Release date: | 2011-05-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of uncharacterized protein Svir_20580 from Saccharomonospora viridis (V71M mutant)
To be Published
|
|
3SG0
 
 | | The crystal structure of an extracellular ligand-binding receptor from Rhodopseudomonas palustris HaA2 | | Descriptor: | BENZOYL-FORMIC ACID, Extracellular ligand-binding receptor | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.201 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3U0H
 
 | | The Structure of a Xylose Isomerase domain protein from Alicyclobacillus acidocaldarius subsp. acidocaldarius. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Cuff, M.E, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-11-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structure of a Xylose Isomerase domain protein from Alicyclobacillus acidocaldarius subsp. acidocaldarius.
TO BE PUBLISHED
|
|
3U24
 
 | | The structure of a putative lipoprotein of unknown function from Shewanella oneidensis. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Cuff, M.E, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-09-30 | | Release date: | 2012-05-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of a putative lipoprotein of unknown function from Shewanella oneidensis.
TO BE PUBLISHED
|
|
3T6O
 
 | | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus. | | Descriptor: | CHLORIDE ION, Sulfate transporter/antisigma-factor antagonist STAS | | Authors: | Cuff, M.E, Moser, C, Hatzos-Skintges, C, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of an Anti-sigma-factor antagonist (STAS) domain protein from Planctomyces limnophilus.
TO BE PUBLISHED
|
|
3SQN
 
 | | Putative Mga family transcriptional regulator from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Putative Mga family transcriptional regulator from Enterococcus faecalis.
To be Published
|
|
3RPD
 
 | | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine. | | Descriptor: | GLYCEROL, Methionine synthase (B12-independent), SELENOMETHIONINE, ... | | Authors: | Cuff, M.E, Li, H, Hatzos-Skintges, C, Tesar, C, Bearden, J, Clancy, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-26 | | Release date: | 2011-08-31 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of a B12-independent methionine synthase from Shewanella sp. W3-18-1 in complex with Selenomethionine.
TO BE PUBLISHED
|
|
3RPW
 
 | | The crystal structure of an ABC transporter from Rhodopseudomonas palustris CGA009 | | Descriptor: | ABC transporter, FORMIC ACID, GLYCEROL, ... | | Authors: | Tan, K, Mack, J.C, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-05-11 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and functional characterization of solute binding proteins for aromatic compounds derived from lignin: p-Coumaric acid and related aromatic acids.
Proteins, 81, 2013
|
|
3RF6
 
 | | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae | | Descriptor: | CITRATE ANION, MAGNESIUM ION, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Nocek, B, Kuznetsova, K, Evdokimova, E, Savchenko, A, Iakunine, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-05 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of glycerol-3 phosphate bound HAD-like phosphatase from Saccharomyces cerevisiae
TO BE PUBLISHED
|
|
3RQB
 
 | |
3RJT
 
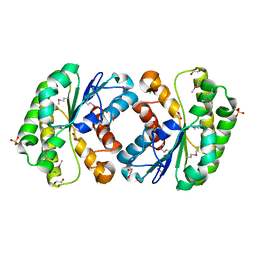 | | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Chang, C, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-15 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of lipolytic protein G-D-S-L family from Alicyclobacillus acidocaldarius subsp. acidocaldarius DSM 446
To be Published
|
|
3RKV
 
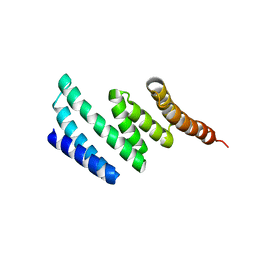 | | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans | | Descriptor: | putative peptidylprolyl isomerase | | Authors: | Osipiuk, J, Tesar, C, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-18 | | Release date: | 2011-05-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | C-terminal domain of protein C56C10.10, a putative peptidylprolyl isomerase, from Caenorhabditis elegans
To be Published
|
|
3RNL
 
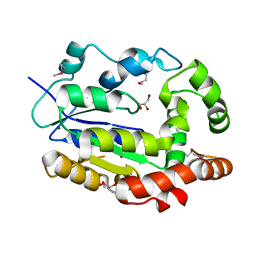 | | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius | | Descriptor: | GLYCEROL, MAGNESIUM ION, Sulfotransferase | | Authors: | Kim, Y, Chhor, G, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-22 | | Release date: | 2011-06-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Crystal Structure of Sulfotransferase from Alicyclobacillus acidocaldarius
To be Published
|
|
3ROB
 
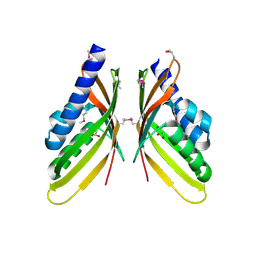 | |
3RQ2
 
 | | Crystal Structure of ADP/ATP-dependent NAD(P)H-hydrate dehydratase from Bacillus subtilis co-crystallized with ATP/Mg2+ and soaked with NADH | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP/ATP-dependent NAD(P)H-hydrate dehydratase, BETA-6-HYDROXY-1,4,5,6-TETRHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ... | | Authors: | Shumilin, I.A, Cymborowski, M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-04-27 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of unknown protein function using metabolite cocktail screening.
Structure, 20, 2012
|
|
3R1X
 
 | | Crystal structure of 2-oxo-3-deoxygalactonate kinase from Klebsiella pneumoniae | | Descriptor: | 2-oxo-3-deoxygalactonate kinase, FORMIC ACID, GLYCEROL | | Authors: | Michalska, K, Cuff, M.E, Tesar, C, Feldmann, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-03-11 | | Release date: | 2011-04-13 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Structure of 2-oxo-3-deoxygalactonate kinase from Klebsiella pneumoniae.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
