3L3G
 
 | | Crystal structure of HLA-B*4402 in complex with the R5A mutant of a self-peptide derived from DPA*0201 | | Descriptor: | ACETATE ION, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Theodossis, A, Ely, L.K, Rossjohn, J. | | Deposit date: | 2009-12-16 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Constraints within major histocompatibility complex class I restricted peptides: presentation and consequences for T-cell recognition
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1CTF
 
 | |
6ZDR
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with Chromone 4d | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Verdon, G, Jespers, W, Azuaje, J, Majellaro, M, Keranen, H, Garcia-mera, X, Congreve, M, Deflorian, F, de Graaf, C, Zhukov, A, Dore, A, Mason, J, Aqvist, J, Cooke, R, Sotelo, E, Gutierrez-de-Teran, H. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | X-Ray Crystallography and Free Energy Calculations Reveal the Binding Mechanism of A 2A Adenosine Receptor Antagonists.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2EFK
 
 | | Crystal structure of the EFC domain of Cdc42-interacting protein 4 | | Descriptor: | Cdc42-interacting protein 4 | | Authors: | Shimada, A, Niwa, H, Chen, L, Liu, Z.-J, Wang, B.-C, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-23 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Curved EFC/F-BAR-Domain Dimers Are Joined End to End into a Filament for Membrane Invagination in Endocytosis
Cell(Cambridge,Mass.), 129, 2007
|
|
4PK3
 
 | | tubulin acetyltransferase complex with bisubstrate analog | | Descriptor: | ACETYL-SER-ASP-(N-ACETYL-LYS)-THR-NH2 PEPTIDE, Alpha-tubulin N-acetyltransferase 1, COENZYME A | | Authors: | Szyk, A, Roll-Mecak, A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.347 Å) | | Cite: | Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase.
Cell, 157, 2014
|
|
4PTM
 
 | | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Madhuprakash, J, Singh, A, Bhushan, A, Sinha, M, Kaur, P, Sharma, S, Podile, A.R, Singh, T.P. | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Chitinase D from Serratia proteamaculans in complex with N-acetyl glucosamine, a hydrolyzed product of hexasaccharide at 1.7 Angstrom resolution
To be Published
|
|
2VG6
 
 | | Crystal structures of HIV-1 reverse transcriptase complexes with thiocarbamate non-nucleoside inhibitors | | Descriptor: | O-[2-(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)ethyl] (4-bromophenyl)thiocarbamate, P51 RT, REVERSE TRANSCRIPTASE/RIBONUCLEASE H | | Authors: | Spallarossa, A, Cesarini, S, Ranise, A, Ponassi, M, Unge, T, Bolognesi, M. | | Deposit date: | 2007-11-08 | | Release date: | 2007-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase Complexes with Thiocarbamate Non-Nucleoside Inhibitors.
Biochem.Biophys.Res.Commun., 365, 2008
|
|
2V2G
 
 | |
7UPZ
 
 | | Structural basis for cell type specific DNA binding of C/EBPbeta: the case of cell cycle inhibitor p15INK4b promoter | | Descriptor: | CCAAT/enhancer-binding protein beta, DNA (5'-D(*AP*TP*TP*CP*TP*TP*AP*AP*GP*AP*AP*AP*GP*AP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*TP*CP*TP*TP*TP*CP*TP*TP*AP*AP*GP*AP*A)-3') | | Authors: | Lountos, G.T, Cherry, S, Tropea, J.E, Wlodawer, A, Miller, M. | | Deposit date: | 2022-04-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.487 Å) | | Cite: | Structural basis for cell type specific DNA binding of C/EBP beta : The case of cell cycle inhibitor p15INK4b promoter.
J.Struct.Biol., 214, 2022
|
|
7UXD
 
 | |
3C2B
 
 | | Crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens | | Descriptor: | FORMIC ACID, Transcriptional regulator, TetR family | | Authors: | Osipiuk, J, Skarina, T, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray crystal structure of TetR transcriptional regulator from Agrobacterium tumefaciens.
To be Published
|
|
3V12
 
 | | Bovine trypsin variant X(tripleGlu217Phe227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
2V41
 
 | |
3JW1
 
 | | Crystal Structure of Bovine Pancreatic Ribonuclease Complexed with Uridine-5'-monophosphate at 1.60 A Resolution | | Descriptor: | Ribonuclease pancreatic, URIDINE-5'-MONOPHOSPHATE | | Authors: | Larson, S.B, Day, J.S, Nguyen, C, Cudney, R, Mcpherson, A, Center for High-Throughput Structural Biology (CHTSB) | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of bovine pancreatic ribonuclease complexed with uridine 5'-monophosphate at 1.60 A resolution.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
6N05
 
 | | Structure of anti-crispr protein, AcrIIC2 | | Descriptor: | AcrIIC2 | | Authors: | Shah, M, Thavalingham, A, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2018-11-06 | | Release date: | 2019-06-05 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition of CRISPR-Cas9 ribonucleoprotein complex assembly by anti-CRISPR AcrIIC2.
Nat Commun, 10, 2019
|
|
7V0J
 
 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellobiose product | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
4RD8
 
 | | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1 | | Descriptor: | Uncharacterized protein | | Authors: | Tan, K, Xu, X, Cui, H, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of a functionally-unknown protein from Legionella pneumophila subsp. pneumophila str. Philadelphia 1
To be Published
|
|
7UNP
 
 | | Crystal structure of the CelR catalytic domain and CBM3c | | Descriptor: | CALCIUM ION, Glucanase | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
4RGJ
 
 | | Apo crystal structure of CDPK4 from Plasmodium falciparum, PF3D7_0717500 | | Descriptor: | Calcium-dependent protein kinase 4 | | Authors: | Wernimont, A.K, Walker, J.R, Hutchinson, A, Seitova, A, He, H, Loppnau, P, Neculai, M, Amani, M, Lin, Y.H, Ravichandran, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Lovato, D.V, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-30 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Apo crystal structure of CDPK4 from Plasmodium falciparum, PF3D7_0717500
To be Published
|
|
7V0I
 
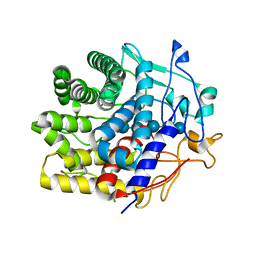 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7V0N
 
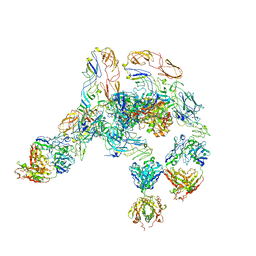 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-21 | | Descriptor: | IgG 21 Fab heavy chain, IgG 21 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3ZMG
 
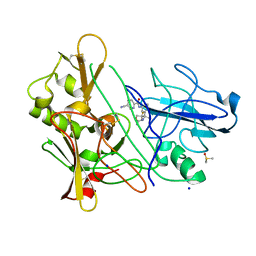 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | BETA-SECRETASE 1, DIMETHYL SULFOXIDE, N-[3-[(4R)-2-azanylidene-5,5-bis(fluoranyl)-4-methyl-1,3-oxazinan-4-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M. | | Deposit date: | 2013-02-08 | | Release date: | 2013-05-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Beta-Secretase (Bace1) Inhibitors with High in Vivo Efficacy Suitable for Clinical Evaluation in Alzheimer'S Disease.
J.Med.Chem., 56, 2013
|
|
7V0O
 
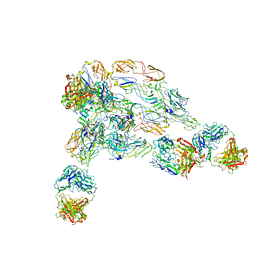 | | Cryo-EM structure of SINV/EEEV in complex with Fab fragment of a moderately/weakly neutralizing human antibody IgG-94 | | Descriptor: | IgG 94 Fab heavy chain, IgG 94 Fab light chain, Spike glycoprotein E1, ... | | Authors: | Bandyopadhyay, A, Klose, T, Kuhn, R.J. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural constraints link differences in neutralization potency of human anti-Eastern equine encephalitis virus monoclonal antibodies.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3KFO
 
 | | Crystal structure of the C-terminal domain from the nuclear pore complex component NUP133 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Nucleoporin NUP133 | | Authors: | Sampathkumar, P, Bonanno, J.B, Miller, S, Bain, K, Dickey, M, Gheyi, T, Almo, S.C, Rout, M, Sali, A, Phillips, J, Pieper, U, Fernandez-Martinez, J, Franke, J.D, Atwell, S, Thompson, D.A, Emtage, J.S, Wasserman, S, Sauder, J.M, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-10-27 | | Release date: | 2010-01-26 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex.
Proteins, 79, 2011
|
|
5EFV
 
 | | The host-recognition device of Staphylococcus aureus phage Phi11 | | Descriptor: | FE (III) ION, Phi ETA orf 56-like protein | | Authors: | Koc, C, Kuhner, P, Xia, G, Spinelli, S, Roussel, A, Cambillau, C, Stehle, T. | | Deposit date: | 2015-10-26 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the host-recognition device of Staphylococcus aureus phage 11.
Sci Rep, 6, 2016
|
|
