3SXZ
 
 | |
3SXM
 
 | |
3SXY
 
 | |
2ETS
 
 | |
2F46
 
 | |
2EVR
 
 | |
2FG0
 
 | |
2FNA
 
 | |
2GAZ
 
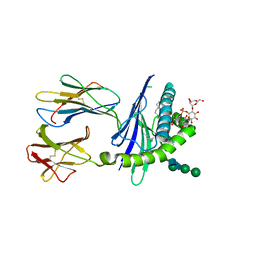 | | Mycobacterial lipoglycan presentation by CD1d | | Descriptor: | (2R)-3-[(HYDROXY{[(2R,3R,5S,6R)-3,4,5-TRIHYDROXY-2,6-BIS(ALPHA-D-MANNOPYRANOSYLOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]PROPAN E-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, T-cell surface glycoprotein CD1d1, ... | | Authors: | Zajonc, D.M. | | Deposit date: | 2006-03-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d.
J.Immunol., 177, 2006
|
|
2H1T
 
 | |
2GHR
 
 | |
2FIK
 
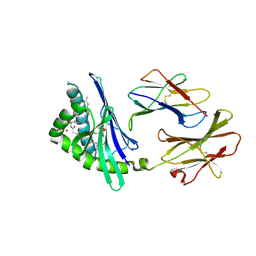 | | Structure of a microbial glycosphingolipid bound to mouse CD1d | | Descriptor: | (2S,3R)-3-HYDROXY-2-(TETRADECANOYLAMINO)OCTADECYL ALPHA-D-GALACTOPYRANOSIDURONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, D, Zajonc, D.M. | | Deposit date: | 2005-12-29 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2G36
 
 | |
2FO4
 
 | |
2FNO
 
 | |
2HBW
 
 | |
2GVI
 
 | |
2HAG
 
 | |
2FEA
 
 | |
2GLZ
 
 | |
2HUJ
 
 | |
2IAY
 
 | |
2GVK
 
 | |
2ICH
 
 | |
2IIZ
 
 | |
