1RHH
 
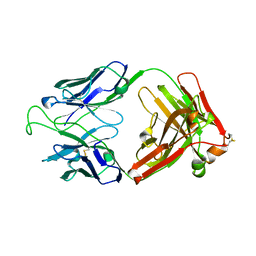 | | Crystal Structure of the Broadly HIV-1 Neutralizing Fab X5 at 1.90 Angstrom Resolution | | Descriptor: | Fab X5, heavy chain, light chain | | Authors: | Darbha, R, Phogat, S, Labrijn, A.F, Shu, Y, Gu, Y, Andrykovitch, M, Zhang, M.Y, Pantophlet, R, Martin, L, Vita, C, Burton, D.R, Dimitrov, D.S, Ji, X. | | Deposit date: | 2003-11-14 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Broadly Cross-Reactive HIV-1-Neutralizing Fab X5 and Fine Mapping of Its Epitope
Biochemistry, 43, 2004
|
|
6BW4
 
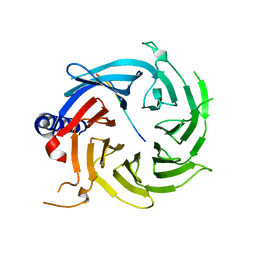 | | Crystal structure of RBBP4 in complex with PRDM16 N-terminal peptide | | Descriptor: | Histone-binding protein RBBP4, PR domain zinc finger protein 16, UNKNOWN ATOM OR ION | | Authors: | Ivanochko, D, Halabelian, L, Hutchinson, A, Seitova, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-12-14 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex.
Nucleic Acids Res., 47, 2019
|
|
6BWM
 
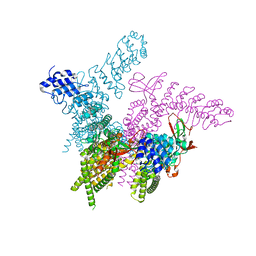 | | Crystal structure of the TRPV2 ion channel | | Descriptor: | CALCIUM ION, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Zubcevic, L, Le, S, Yang, H, Lee, S.Y. | | Deposit date: | 2017-12-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Conformational plasticity in the selectivity filter of the TRPV2 ion channel.
Nat. Struct. Mol. Biol., 25, 2018
|
|
1YGH
 
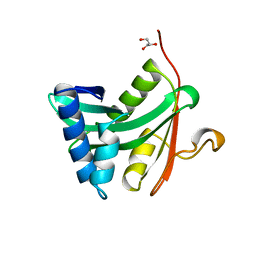 | | HAT DOMAIN OF GCN5 FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | GLYCEROL, PROTEIN (TRANSCRIPTIONAL ACTIVATOR GCN5) | | Authors: | Trievel, R.C, Rojas, J.R, Sterner, D.E, Venkataramani, R, Wang, L, Zhou, J, Allis, C.D, Berger, S.L, Marmorstein, R. | | Deposit date: | 1999-05-27 | | Release date: | 1999-08-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5EKE
 
 | | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB (F215A mutant) | | Descriptor: | MAGNESIUM ION, URIDINE-5'-DIPHOSPHATE, Uncharacterized glycosyltransferase sll0501 | | Authors: | Ardiccioni, C, Clarke, O.B, Tomasek, D, Banerjee, S, Rajashankar, K.R, Liu, Q, Shapiro, L, Mancia, F, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2015-11-03 | | Release date: | 2016-01-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Structure of the polyisoprenyl-phosphate glycosyltransferase GtrB and insights into the mechanism of catalysis.
Nat Commun, 7, 2016
|
|
1Y89
 
 | | Crystal Structure of devB protein | | Descriptor: | 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, DI(HYDROXYETHYL)ETHER, NONAETHYLENE GLYCOL, ... | | Authors: | Lazarski, K, Cymborowski, M, Chruszcz, M, Zheng, H, Zhang, R, Lezondra, L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of devB protein
To be Published
|
|
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C0M
 
 | | The synthesis, biological evaluation and structural insights of unsaturated 3-N-substituted sialic acids as probes of human parainfluenza virus-3 haemagglutinin-neuraminidase | | Descriptor: | 1,2-ETHANEDIOL, 2,6-anhydro-3,5-dideoxy-5-[(2-methylpropanoyl)amino]-3-(4-phenyl-1H-1,2,3-triazol-1-yl)-D-glycero-D-galacto-non-2-enoni c acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dirr, L, Ve, T, von Itzstein, M. | | Deposit date: | 2018-01-01 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insights into Human Parainfluenza Virus 3 Hemagglutinin-Neuraminidase Using Unsaturated 3- N-Substituted Sialic Acids as Probes.
ACS Chem. Biol., 13, 2018
|
|
5EMK
 
 | |
1YB5
 
 | | Crystal structure of human Zeta-Crystallin with bound NADP | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Debreczeni, J, Berridge, G, Kavanagh, K, Colbrook, S, Bray, J, Williams, L, Oppermann, U, Sundstrom, M, Arrowsmith, C, Edwards, A, Gileadi, O, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2004-12-20 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of human Zeta-Crystallin at 1.85A
To be Published
|
|
5EO3
 
 | |
1YDM
 
 | | X-Ray structure of Northeast Structural Genomics target SR44 | | Descriptor: | Hypothetical protein yqgN, SULFATE ION | | Authors: | Kuzin, A.P, Jianwei, S, Vorobiev, S, Acton, T, Xia, R, Ma, L.-C, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-24 | | Release date: | 2005-01-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray structure of Northeast Structural Genomics target SR44
To be Published
|
|
1RQB
 
 | | Propionibacterium shermanii transcarboxylase 5S subunit | | Descriptor: | COBALT (II) ION, transcarboxylase 5S subunit | | Authors: | Hall, P.R, Zheng, R, Antony, L, Pusztai-Carey, M, Carey, P.R, Yee, V.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-09-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Transcarboxylase 5S structures: assembly and catalytic mechanism of a multienzyme complex subunit.
Embo J., 23, 2004
|
|
1YGG
 
 | | Crystal structure of phosphoenolpyruvate carboxykinase from Actinobacillus succinogenes | | Descriptor: | SODIUM ION, SULFATE ION, phosphoenolpyruvate carboxykinase | | Authors: | Leduc, Y.A, Prasad, L, Laivenieks, M, Zeikus, J.G, Delbaere, L.T. | | Deposit date: | 2005-01-04 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PEP carboxykinase from the succinate-producing Actinobacillus succinogenes: a new conserved active-site motif.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6C5N
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
6C7A
 
 | | Conformational Changes in a Class A Beta lactamase that Prime it for Catalysis | | Descriptor: | Beta-lactamase Toho-1, SULFATE ION | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Cooper, S.J, Weiss, K.L, Ginell, S.L, Parks, J.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate Binding Induces Conformational Changes in a Class A Beta-lactamase That Prime It for Catalysis
Acs Catalysis, 8, 2018
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
1Y7I
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | 2-HYDROXYBENZOIC ACID, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6C4O
 
 | |
5EAZ
 
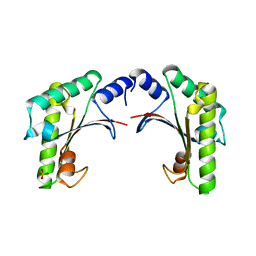 | | crystal form I of YfiB belonging to space groups P21 | | Descriptor: | SULFATE ION, YfiB | | Authors: | Xu, M, Yang, X, Yang, X.-A, Zhou, L, Liu, T.-Z, Fan, Z, Jiang, T. | | Deposit date: | 2015-10-17 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Structural insights into the regulatory mechanism of the Pseudomonas aeruginosa YfiBNR system
Protein Cell, 7, 2016
|
|
5DQP
 
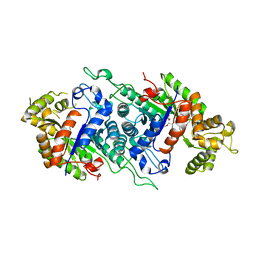 | | EDTA monooxygenase (EmoA) from Chelativorans sp. BNC1 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, EDTA monooxygenase, SULFATE ION | | Authors: | Jun, S.Y, Youn, B, Xun, L, Kang, C, Lewis, K.M. | | Deposit date: | 2015-09-15 | | Release date: | 2016-03-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.146 Å) | | Cite: | Structural and biochemical characterization of EDTA monooxygenase and its physical interaction with a partner flavin reductase.
Mol.Microbiol., 100, 2016
|
|
1YJX
 
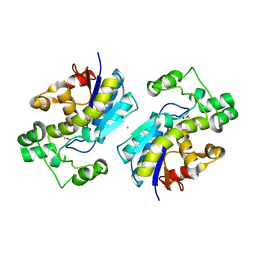 | | Crystal structure of human B type phosphoglycerate mutase | | Descriptor: | CHLORIDE ION, CITRIC ACID, Phosphoglycerate mutase 1 | | Authors: | Wang, Y, Wei, Z, Liu, L, Gong, W. | | Deposit date: | 2005-01-16 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human B-type phosphoglycerate mutase bound with citrate.
Biochem.Biophys.Res.Commun., 331, 2005
|
|
1YJU
 
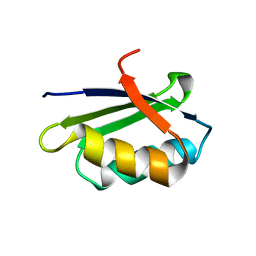 | | Solution structure of the apo form of the sixth soluble domain of Menkes protein | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Cantini, F, Migliardi, M, Rosato, A, Wang, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-01-15 | | Release date: | 2006-01-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | An atomic-level investigation of the disease-causing A629P mutant of the Menkes protein, ATP7A
J.Mol.Biol., 352, 2005
|
|
1US3
 
 | | Native xylanase10C from Cellvibrio japonicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-1,4-XYLANASE PRECURSOR, GLYCEROL, ... | | Authors: | Pell, G, Szabo, L, Charnock, S.J, Xie, H, Gloster, T.M, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2003-11-17 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Analysis of Cellvibrio Japonicus Xylanase 10C: How Variation in Substrate-Binding Cleft Influences the Catalytic Profile of Family Gh-10 Xylanases
J.Biol.Chem., 279, 2004
|
|
