3BPK
 
 | | Crystal structure of nitrilotriacetate monooxygenase component B from Bacillus cereus | | Descriptor: | CHLORIDE ION, Nitrilotriacetate monooxygenase component B, SULFATE ION | | Authors: | Osipiuk, J, Quartey, P, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-18 | | Release date: | 2008-01-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | X-ray crystal structure of nitrilotriacetate monooxygenase component B from Bacillus cereus.
To be Published
|
|
3GHJ
 
 | | Crystal structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4 | | Descriptor: | Putative integron gene cassette protein | | Authors: | Sureshan, V, Deshpande, C, Harrop, S.J, Kudritska, M, Koenig, J.E, Evdokimova, E, Kim, Y, Edwards, A.M, Savchenko, A, Joachimiak, A, Doolittle, W.F, Stokes, H.W, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-03 | | Release date: | 2009-03-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.471 Å) | | Cite: | Structure from the mobile metagenome of Halifax Harbour Sewage Outfall: Integron Cassette Protein HFX_CASS4
To be Published
|
|
3NET
 
 | |
3GKX
 
 | |
3MWB
 
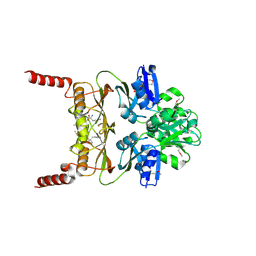 | | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A | | Descriptor: | MAGNESIUM ION, PHENYLALANINE, Prephenate dehydratase | | Authors: | Stein, A.J, Chhor, G, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Prephenate dehydratase in complex with L-Phe from Arthrobacter aurescens to 2.0A
To be Published
|
|
3C8D
 
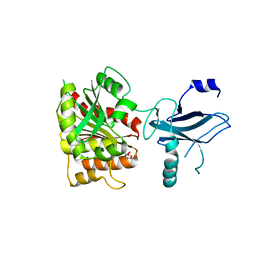 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-glycine | | Descriptor: | CITRIC ACID, Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-11 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
3JTF
 
 | | The CBS Domain Pair Structure of a magnesium and cobalt efflux protein from Bordetella parapertussis in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Magnesium and cobalt efflux protein, SULFATE ION | | Authors: | Cuff, M.E, Tesar, C, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-11 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The CBS Domain Pair Structure of a magnesium and cobalt efflux protein from Bordetella parapertussis in complex with AMP
TO BE PUBLISHED
|
|
3CDH
 
 | | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi DSS-3 | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Kim, Y, Volkart, L, Keigher, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-26 | | Release date: | 2008-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of the MarR family transcriptional regulator SPO1453 from Silicibacter pomeroyi.
To be Published
|
|
3JU1
 
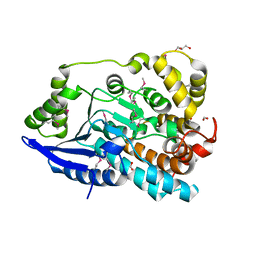 | | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein | | Descriptor: | ACETIC ACID, Enoyl-CoA hydratase/isomerase family protein, FORMIC ACID, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-14 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Crystal Structure of Enoyl-CoA Hydratase/Isomerase Family Protein
To be Published
|
|
3JUW
 
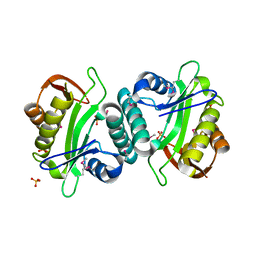 | |
3MUQ
 
 | |
3K2V
 
 | | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae. | | Descriptor: | CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL, SULFATE ION, ... | | Authors: | Cuff, M.E, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-30 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Klebsiella pneumoniae subsp. pneumoniae.
TO BE PUBLISHED
|
|
3K6C
 
 | | Crystal structure of protein ne0167 from nitrosomonas europaea | | Descriptor: | Uncharacterized protein NE0167 | | Authors: | Chang, C, Evdokimova, E, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-08 | | Release date: | 2009-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Protein Ne0167 from Nitrosomonas Europaea
To be Published
|
|
1YB4
 
 | | Crystal Structure of the Tartronic Semialdehyde Reductase from Salmonella typhimurium LT2 | | Descriptor: | tartronic semialdehyde reductase | | Authors: | Kim, Y, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-20 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystal structure of GarR-tartronate semialdehyde reductase from Salmonella typhimurium.
J Struct Funct Genomics, 10, 2009
|
|
3NZN
 
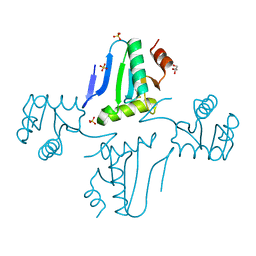 | | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1 | | Descriptor: | GLYCEROL, Glutaredoxin, SULFATE ION | | Authors: | Zhang, R, Wu, R, Freeman, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-16 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The crystal structure of the Glutaredoxin from Methanosarcina mazei Go1
To be Published
|
|
3HCY
 
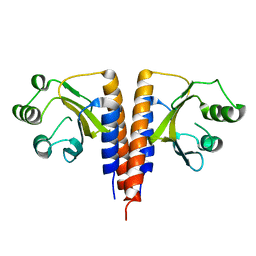 | | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021 | | Descriptor: | Putative two-component sensor histidine kinase protein | | Authors: | Zhang, R, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of the domain of putative two-component sensor histidine kinase protein from Sinorhizobium meliloti 1021
To be Published
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN LIGAND | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
3CK2
 
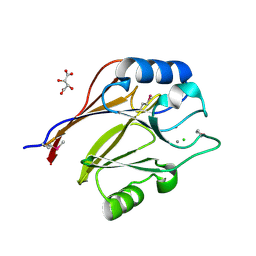 | | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4 | | Descriptor: | CHLORIDE ION, Conserved uncharacterized protein (predicted phosphoesterase COG0622), MANGANESE (II) ION, ... | | Authors: | Nocek, B, Zhou, M, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of conserved uncharacterized protein (predicted phosphoesterase COG0622) from Streptococcus pneumoniae TIGR4.
To be Published
|
|
3CKD
 
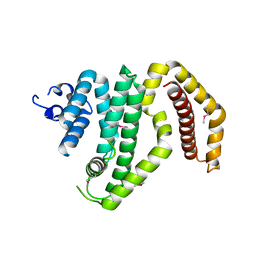 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3O5Y
 
 | |
4R1H
 
 | |
3D6J
 
 | | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative haloacid dehalogenase-like hydrolase | | Authors: | Chang, C, Wu, R, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-19 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Putative haloacid dehalogenase-like hydrolase from Bacteroides fragilis
To be Published
|
|
3JRK
 
 | | A putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes | | Descriptor: | GLYCEROL, Tagatose 1,6-diphosphate aldolase 2 | | Authors: | Fan, Y, Volkart, L, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The crystal structure of a putative tagatose 1,6-diphosphate aldolase from Streptococcus pyogenes
To be Published
|
|
3JW4
 
 | | The structure of a putative MarR family transcriptional regulator from Clostridium acetobutylicum | | Descriptor: | GLYCEROL, IMIDAZOLE, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Bigelow, L, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a putative MarR family transcriptional regulator from Clostridium acetobutylicum
TO BE PUBLISHED
|
|
3BQW
 
 | |
