7MEL
 
 | | Sco GlgEI-V279S in complex with 4-alpha-glucoside of validamine | | Descriptor: | (1R,2R,3S,4S,6R)-4-amino-2,3-dihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-04-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereoselective synthesis of a 4-alpha-glucoside of valienamine and its X-ray structure in complex with Streptomyces coelicolor GlgE1-V279S.
Sci Rep, 11, 2021
|
|
7MGY
 
 | | Sco GlgEI-V279S in complex with 4-alpha-glucoside of valienamine | | Descriptor: | (1R,4S,5S,6R)-4-amino-5,6-dihydroxy-2-(hydroxymethyl)cyclohex-2-en-1-yl alpha-D-glucopyranoside, Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jayasinghe, T.D, Ronning, D.R. | | Deposit date: | 2021-04-13 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Stereoselective synthesis of a 4-⍺-glucoside of valienamine and its X-ray structure in complex with Streptomyces coelicolor GlgE1-V279S.
Sci Rep, 11, 2021
|
|
6IWI
 
 | | Crystal structure of PDE5A in complex with a novel inhibitor | | Descriptor: | MAGNESIUM ION, N-[3-(4,5-diethyl-6-oxo-1,6-dihydropyrimidin-2-yl)-4-propoxyphenyl]-2-(4-methylpiperazin-1-yl)acetamide, ZINC ION, ... | | Authors: | Zhang, X.L, Xu, Y.C. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Pharmacokinetics-Driven Optimization of 4(3 H)-Pyrimidinones as Phosphodiesterase Type 5 Inhibitors Leading to TPN171, a Clinical Candidate for the Treatment of Pulmonary Arterial Hypertension.
J.Med.Chem., 62, 2019
|
|
4QRS
 
 | | Crystal Structure of HLA B*0801 in complex with ELK_IYM, ELKRKMIYM | | Descriptor: | ACETATE ION, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Twist, K.-A, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-12-10 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular imprint of exposure to naturally occurring genetic variants of human cytomegalovirus on the T cell repertoire.
Sci Rep, 4, 2014
|
|
5V85
 
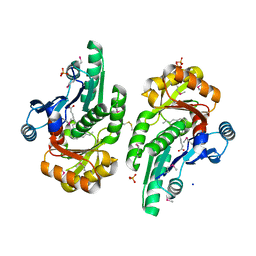 | | The crystal structure of the protein of DegV family COG1307 from Ruminococcus gnavus ATCC 29149 (alternative refinement of PDB 3JR7 with Vaccenic acid) | | Descriptor: | EDD domain protein, DegV family, PHOSPHATE ION, ... | | Authors: | Cuypers, M.G, Ericson, M, subramanian, C, White, S.W, Rock, C.O. | | Deposit date: | 2017-03-21 | | Release date: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | The crystal structure of the Staphylococcus aureus Fatty acid Kinase (Fak) B1 protein loaded with palmitic acid to 1.83 Angstroem resolution
J.Biol.Chem., 2018
|
|
8H0F
 
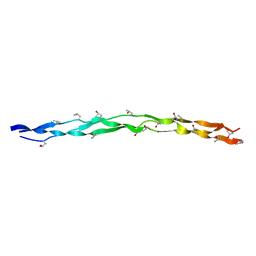 | | Crystal structure of collagen heterotrimer with KD,ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
8GZO
 
 | |
8H0E
 
 | | Crystal structure of collagen heterotrimer with KD, ER and KE axial pairs | | Descriptor: | collagen-like peptide chain A, collagen-like peptide chain B, collagen-like peptide chain C | | Authors: | Fan, S. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Stability of collagen heterotrimer with same charge pattern and different charged residue identities.
Biophys.J., 122, 2023
|
|
7E8M
 
 | | Crystal structure of SARS-CoV-2 antibody P2C-1F11 with mutated RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody P2C-1F11 heavy chain, ... | | Authors: | Wang, X.Q, Zhang, L.Q, Ge, J.W, Wang, R.K, Lan, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species.
Immunity, 54, 2021
|
|
5HLT
 
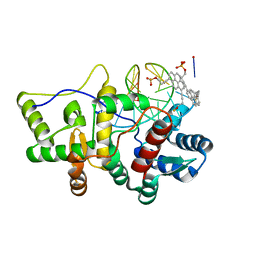 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*YPY*TP*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*AP*YPY*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.672 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
7E7E
 
 | | The co-crystal structure of ACE2 with Fab | | Descriptor: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
5Z9J
 
 | | Identification of the functions of unusual cytochrome p450-like monooxygenases involved in microbial secondary metablism | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Putative P450-like enzyme, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Lu, M, Lin, L, Zhang, C, Chen, Y. | | Deposit date: | 2018-02-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Riboflavin Is Directly Involved in the N-Dealkylation Catalyzed by Bacterial Cytochrome P450 Monooxygenases.
Chembiochem, 2020
|
|
6C8U
 
 | | Solution structure of Musashi2 RRM1 | | Descriptor: | RNA-binding protein Musashi homolog 2 | | Authors: | Xing, M, Lan, L, Douglas, J.T, Gao, P, Hanzlik, R.P, Xu, L. | | Deposit date: | 2018-01-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 2019
|
|
6NTY
 
 | | 2.1 A resolution structure of the Musashi-2 (Msi2) RNA recognition motif 1 (RRM1) domain | | Descriptor: | PHOSPHATE ION, RNA-binding protein Musashi homolog 2 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Lan, L, Xiaoqing, W, Cooper, A, Gao, F.P, Xu, L. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal and solution structures of human oncoprotein Musashi-2 N-terminal RNA recognition motif 1.
Proteins, 88, 2020
|
|
7FJO
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with three T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
5Z9I
 
 | |
2G9T
 
 | | Crystal structure of the SARS coronavirus nsp10 at 2.1A | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Yang, H, Sun, F, Rao, Z. | | Deposit date: | 2006-03-07 | | Release date: | 2006-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
7FJN
 
 | | Cryo-EM structure of South African (B.1.351) SARS-CoV-2 spike glycoprotein in complex with two T6 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, T6 heavy chain, ... | | Authors: | Wang, X, Zhang, L, Zhang, S, Liang, Q. | | Deposit date: | 2021-08-04 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | RBD trimer mRNA vaccine elicits broad and protective immune responses against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
6NLE
 
 | | X-ray structure of LeuT with V269 deletion | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Navratna, V, Yang, D, Gouaux, E. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.615 Å) | | Cite: | Structural, functional, and behavioral insights of dopamine dysfunction revealed by a deletion inSLC6A3.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4GH4
 
 | | Crystal Structure of Foot and Mouth Disease Virus A22 Serotype | | Descriptor: | capsid protein VP1, capsid protein VP2, capsid protein VP3, ... | | Authors: | Kotecha, A, Jinshan, R, Curry, S, Fry, E, Stuart, D. | | Deposit date: | 2012-08-07 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Perturbations in the surface structure of A22 Iraq foot-and-mouth disease virus accompanying coupled changes in host cell specificity and antigenicity.
Structure, 4, 1996
|
|
5HNH
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*(YPY)P*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*T*(YPY)*(YPY)*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.876 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
1KR4
 
 | | Structure Genomics, Protein TM1056, cutA | | Descriptor: | Protein TM1056, cutA | | Authors: | Savchenko, A, Zhang, R, Joachimiak, A, Edwards, A, Akarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-08 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray crystal structure of CutA from Thermotoga maritima at 1.4 A resolution.
Proteins, 54, 2004
|
|
5HNF
 
 | | Crystal structure of pyrene- and phenanthrene-modified DNA in complex with the BpuJ1 endonuclease binding domain | | Descriptor: | DNA (5'-D(*GP*(YPE)P*AP*CP*CP*CP*GP*TP*GP*GP*A)-3'), DNA (5'-D(*TP*CP*CP*AP*CP*GP*GP*GP*TP*(YPF)P*C)-3'), Restriction endonuclease R.BpuJI | | Authors: | Probst, M, Aeschimann, W, Chau, T.-T.-H, Langenegger, S.M, Stocker, A, Haener, R. | | Deposit date: | 2016-01-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.546 Å) | | Cite: | Structural insight into DNA-assembled oligochromophores: crystallographic analysis of pyrene- and phenanthrene-modified DNA in complex with BpuJI endonuclease.
Nucleic Acids Res., 44, 2016
|
|
2X2I
 
 | | Crystal structure of the Gracilariopsis lemaneiformis alpha-1,4- glucan lyase with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA-1,4-GLUCAN LYASE ISOZYME 1, GLYCEROL | | Authors: | Rozeboom, H.J, Yu, S, Madrid, S, Kalk, K.H, Dijkstra, B.W. | | Deposit date: | 2010-01-13 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Alpha-1,4-Glucan Lyase, a Unique Glycoside Hydrolase Family Member with a Novel Catalytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
2GA6
 
 | | The crystal structure of SARS nsp10 without zinc ion as additive | | Descriptor: | ZINC ION, orf1a polyprotein | | Authors: | Su, D, Lou, Z, Sun, F, Zhai, Y, Yang, H, Rao, Z. | | Deposit date: | 2006-03-08 | | Release date: | 2006-08-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dodecamer Structure of Severe Acute Respiratory Syndrome Coronavirus Nonstructural Protein nsp10
J.Virol., 80, 2006
|
|
