3TSD
 
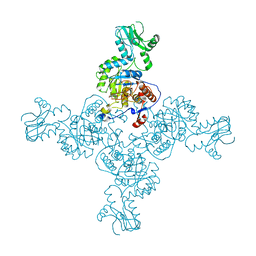 | | Crystal Structure of Inosine-5'-monophosphate Dehydrogenase from Bacillus anthracis str. Ames complexed with XMP | | Descriptor: | D(-)-TARTARIC ACID, Inosine-5'-monophosphate dehydrogenase, SULFATE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-13 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Bacillus anthracis inosine 5'-monophosphate dehydrogenase in action: the first bacterial series of structures of phosphate ion-, substrate-, and product-bound complexes.
Biochemistry, 51, 2012
|
|
4H1H
 
 | |
3UHP
 
 | | Crystal Structure of Glutamate Racemase from Campylobacter jejuni subsp. jejuni | | Descriptor: | Glutamate racemase | | Authors: | Maltseva, N, Mulligan, R, Kwon, K, Kim, Y, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-11-03 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Crystal Structure of Glutamate Racemase
from Campylobacter jejuni subsp. jejuni
To be Published, 2011
|
|
4HTQ
 
 | | Mitigation of X-ray damage in macromolecular crystallography by submicrometer line focusing; total dose 6.70 x 10e+11 X-ray photons | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Duke, N.E.C, Finfrock, Y.Z, Stern, E.Z, Alkire, R.W, Lazarski, K, Joachimiak, A. | | Deposit date: | 2012-11-01 | | Release date: | 2013-05-15 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Mitigation of X-ray damage in macromolecular crystallography by submicrometre line focusing.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TY6
 
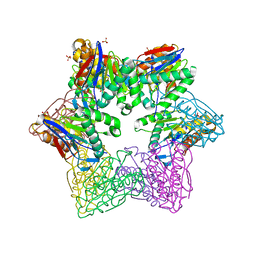 | | ATP-dependent Protease HslV from Bacillus anthracis str. Ames | | Descriptor: | ATP-dependent protease subunit HslV, SULFATE ION | | Authors: | Kim, Y, Mulligan, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-23 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | ATP-dependent Protease HslV from Bacillus anthracis str. Ames
To be Published
|
|
3TJ7
 
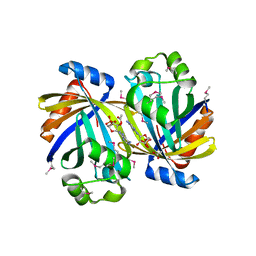 | | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis in complex with AMP | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, GBAA_1210 protein, ... | | Authors: | Osipiuk, J, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-08-23 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GBAA_1210 protein, a putative adenylate cyclase, from Bacillus anthracis in complex with AMP
To be Published
|
|
3TV9
 
 | | Crystal Structure of Putative Peptide Maturation Protein from Shigella flexneri | | Descriptor: | GLYCEROL, Putative peptide maturation protein, SULFATE ION | | Authors: | Kim, Y, Maltseva, N, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-19 | | Release date: | 2011-10-05 | | Last modified: | 2016-12-28 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Crystal Structure of Putative Peptide Maturation Protein from
To be Published
|
|
6B4O
 
 | | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Glutathione reductase, ... | | Authors: | Minasov, G, Warwzak, Z, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-09-27 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | 1.73 Angstrom Resolution Crystal Structure of Glutathione Reductase from Enterococcus faecalis in Complex with FAD.
To Be Published
|
|
1KS2
 
 | | Crystal Structure Analysis of the rpiA, Structural Genomics, protein EC1268. | | Descriptor: | protein EC1268, RPIA | | Authors: | Zhang, R, Joachimiak, A, Edwards, A.M, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-10 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Escherichia coli ribose-5-phosphate isomerase: a ubiquitous enzyme of the pentose phosphate pathway and the Calvin cycle.
STRUCTURE, 11, 2003
|
|
1L8R
 
 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
6B8W
 
 | | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae. | | Descriptor: | MANGANESE (II) ION, THIOCYANATE ION, XRE family transcriptional regulator | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, McChesney, C, Grimshaw, S, Sandoval, J, Satchell, K.J.F, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-10-09 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Cupin_2 Domain (pfam 07883) of XRE Family Transcriptional Regulator from Enterobacter cloacae.
To Be Published
|
|
6CN0
 
 | | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis | | Descriptor: | 16S rRNA (guanine(1405)-N(7))-methyltransferase, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Minasov, G, Wawrzak, Z, Di Leo, R, Evdokimova, E, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | 2.95 Angstrom Crystal Structure of 16S rRNA Methylase from Proteus mirabilis.
To Be Published
|
|
6CN1
 
 | | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-03-06 | | Release date: | 2018-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Pseudomonas putida in Complex with Uridine-diphosphate-2(n-acetylglucosaminyl) butyric acid, (2R)-2-(phosphonooxy)propanoic acid and Magnesium.
To Be Published
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
6CZP
 
 | | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Minasov, G, Wawrzak, Z, Skarina, T, Grimshaw, S, Kwon, K, Savchenko, A, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | 2.2 Angstrom Resolution Crystal Structure Oxygen-Insensitive NAD(P)H-dependent Nitroreductase NfsB from Vibrio vulnificus in Complex with FMN.
To Be Published
|
|
1NE2
 
 | | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513) | | Descriptor: | FORMIC ACID, hypothetical protein ta1320 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513)
To be Published
|
|
6BK7
 
 | | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis | | Descriptor: | Elongation factor G, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Cardona-Correa, A, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-07 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 1.83 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-404) of Elongation Factor G from Enterococcus faecalis.
To be Published
|
|
1KJN
 
 | |
1L1S
 
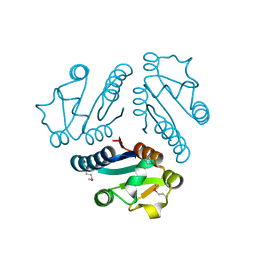 | | Structure of Protein of Unknown Function MTH1491 from Methanobacterium thermoautotrophicum | | Descriptor: | hypothetical protein MTH1491 | | Authors: | Christendat, D, Saridakis, V, Kim, Y, Kumar, P.A, Xu, X, Semesi, A, Joachimiak, A, Arrowsmith, C.H, Edwards, A.M, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of hypothetical protein MTH1491 from Methanobacterium thermoautotrophicum.
Protein Sci., 11, 2002
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1KYT
 
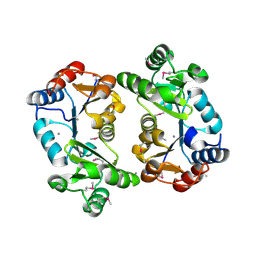 | | Crystal Structure of Thermoplasma acidophilum 0175 (APC014) | | Descriptor: | CALCIUM ION, hypothetical protein TA0175 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Pennycooke, M, Gu, J, Cheung, F, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-02-05 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 0175 (APC014)
To be published
|
|
1L0Q
 
 | | Tandem YVTN beta-propeller and PKD domains from an archaeal surface layer protein | | Descriptor: | Surface layer protein | | Authors: | Jing, H, Takagi, J, Liu, J.-H, Lindgren, S, Zhang, R.-G, Joachimiak, A, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-02-12 | | Release date: | 2002-11-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Archaeal Surface Layer Proteins Contain beta Propeller, PKD, and beta Helix Domains and Are Related to Metazoan Cell Surface Proteins.
Structure, 10, 2002
|
|
1NN4
 
 | | Structural Genomics, RpiB/AlsB | | Descriptor: | Ribose 5-phosphate isomerase B | | Authors: | Zhang, R.G, Andersson, C.E, Mowbray, S.L, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S.L, Arrowsmith, C, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-12 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 A resolution structure of RpiB/AlsB from Escherichia coli illustrates a new approach to the ribose-5-phosphate isomerase reaction.
J.Mol.Biol., 332, 2003
|
|
1MK4
 
 | | Structure of Protein of Unknown Function YqjY from Bacillus subtilis, Probable Acetyltransferase | | Descriptor: | Hypothetical protein yqjY | | Authors: | Zhang, R, Dementiva, I, Mo, A, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-08-28 | | Release date: | 2003-04-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7A crystal structure of a hypothetical protein yqjY
from Bacillus subtilis
To be Published
|
|
1O8B
 
 | | Structure of Escherichia coli ribose-5-phosphate isomerase, RpiA, complexed with arabinose-5-phosphate. | | Descriptor: | 5-O-phosphono-beta-D-arabinofuranose, RIBOSE 5-PHOSPHATE ISOMERASE | | Authors: | Zhang, R.-g, Andersson, C.E, Savchenko, A, Skarina, T, Evdokimova, E, Beasley, S, Arrowsmith, C.H, Edwards, A.M, Joachimiak, A, Mowbray, S.L, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-11-26 | | Release date: | 2003-01-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure of Escherichia Coli Ribose-5-Phosphate Isomerase: A Ubiquitous Enzyme of the Pentose Phosphate Pathway and the Calvin Cycle
Structure, 11, 2003
|
|
