3FK3
 
 | | Structure of the Yeats Domain, Yaf9 | | Descriptor: | Protein AF-9 homolog | | Authors: | Wang, A.Y, Schulze, J.M, Skordalakes, E, Berger, J.M, Rine, J, Kobor, M.S. | | Deposit date: | 2008-12-15 | | Release date: | 2009-10-27 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3IWZ
 
 | | The c-di-GMP Responsive Global Regulator CLP Links Cell-Cell Signaling to Virulence Gene Expression in Xanthomonas campestris | | Descriptor: | Catabolite activation-like protein | | Authors: | Chin, K.H, Tu, Z.L, Tseng, Y.H, Dow, J.M, Wang, A.H.J, Chou, S.H. | | Deposit date: | 2009-09-03 | | Release date: | 2009-12-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris.
J.Mol.Biol., 396, 2010
|
|
3S88
 
 | | Crystal structure of Sudan Ebolavirus Glycoprotein (strain Gulu) bound to 16F6 | | Descriptor: | 16F6 - Heavy chain, 16F6 - Light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Saphire, E.O, Dias, J.M, Bale, S. | | Deposit date: | 2011-05-27 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.351 Å) | | Cite: | A shared structural solution for neutralizing ebolaviruses.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3RTP
 
 | | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration | | Descriptor: | Mitogen-activated protein kinase 10, N-[4-cyano-3-(1H-1,2,4-triazol-5-yl)thiophen-2-yl]-2-(2-oxo-3,4-dihydroquinolin-1(2H)-yl)acetamide | | Authors: | Bowers, S, Truong, A.P, Neitz, R.J, Hom, R.K, Sealy, J.M, Probst, G.D, Quincy, Q, Peterson, B, Chan, W, Galemmo Jr, R.A, Konradi, A.W, Sham, H.L, Pan, H, Lin, M, Yao, N, Artis, D.R, Zhang, H, Chen, L, Dryer, M, Samant, B, Zmolek, W, Wong, K, Lorentzen, C, Goldbach, E, Tonn, G, Quinn, K.P, Sauer, J, Wright, S, Powell, K, Ruslim, L, Ren, Z, Bard, F, Yednock, T.A, Griswold-Prenne, I. | | Deposit date: | 2011-05-03 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of brain penetrant selective JNK inhibitors with improved pharmacokinetic properties for the prevention of neurodegeneration.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1POV
 
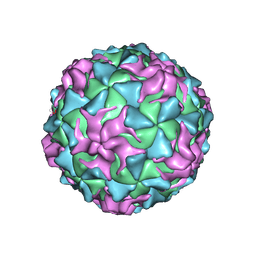 | |
1PND
 
 | |
3G7C
 
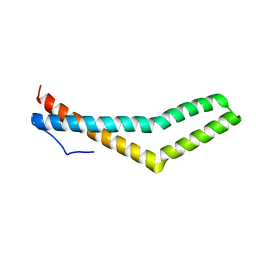 | | Structure of the Phosphorylation Mimetic of Occludin C-term Tail | | Descriptor: | Occludin | | Authors: | Tash, B.R, Sundstrom, J.M, Murakami, T, Flanagan, J.M, Bewley, M.C, Antonetii, D.A. | | Deposit date: | 2009-02-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification and analysis of occludin phosphosites: a combined mass spectrometry and bioinformatics approach.
J.PROTEOME RES., 8, 2009
|
|
1PYN
 
 | | DUAL-SITE POTENT, SELECTIVE PROTEIN TYROSINE PHOSPHATASE 1B INHIBITOR USING A LINKED FRAGMENT STRATEGY AND A MALONATE HEAD ON THE FIRST SITE | | Descriptor: | 2-(4-{2-TERT-BUTOXYCARBONYLAMINO-2-[4-(3-HYDROXY-2-METHOXYCARBONYL-PHENOXY)-BUTYLCARBAMOYL]-ETHYL}-PHENOXY)-MALONIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Szczepankiewicz, B.G, Liu, G, Hajduk, P.J, Abad-Zapatero, C, Zhonghua, P, Lubben, T, Trevillyan, J.M, Stashko, M, Ballaron, S.J, Liang, H. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and SAR of novel, potent and selective protein tyrosine phosphatase 1B inhibitors.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1PNC
 
 | |
3HJ6
 
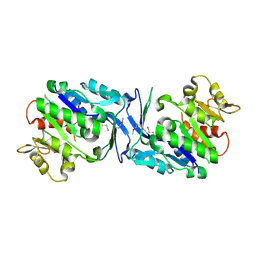 | | Structure of Halothermothrix orenii fructokinase (FRK) | | Descriptor: | Fructokinase | | Authors: | Chua, T.K, Seetharaman, J, Kasprzak, J.M, Ng, C, Patel, B.K, Love, C, Bujnicki, J.M, Sivaraman, J. | | Deposit date: | 2009-05-21 | | Release date: | 2010-06-09 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a fructokinase homolog from Halothermothrix orenii
J.Struct.Biol., 171, 2010
|
|
2IJQ
 
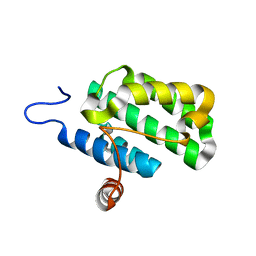 | | Crystal structure of protein rrnAC1037 from Haloarcula marismortui, Pfam DUF309 | | Descriptor: | Hypothetical protein | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Schwinn, K.D, Thompson, D.A, Rutter, M.E, Dickey, M, Groshong, C, Bain, K.T, Adams, J.M, Reyes, C, Rooney, I, Powell, A, Boice, A, Gheyi, T, Ozyurt, S, Atwell, S, Wasserman, S.R, Emtage, S, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-30 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of the hypothetical Protein from Haloarcula marismortui
To be Published
|
|
2IJ9
 
 | | Crystal Structure of Uridylate Kinase from Archaeoglobus Fulgidus | | Descriptor: | SULFATE ION, Uridylate kinase | | Authors: | Patskovsky, Y, Chen, R, Keefe, L.J, Sauder, J.M, Dickey, M, Adams, J.M, Ozyurt, S, Wasserman, S.R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-09-29 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Uridylate Kinase from Archaeoglobus Fulgidus
To be Published
|
|
1JBP
 
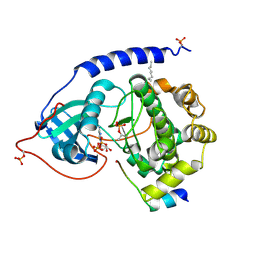 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Substrate Peptide, ADP and Detergent | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, MUSCLE/BRAIN FORM, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-06-06 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
1ATP
 
 | | 2.2 angstrom refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MNATP and a peptide inhibitor | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, PEPTIDE INHIBITOR PKI(5-24), ... | | Authors: | Zheng, J, Trafny, E.A, Knighton, D.R, Xuong, N.-H, Taylor, S.S, Teneyck, L.F, Sowadski, J.M. | | Deposit date: | 1993-01-08 | | Release date: | 1993-04-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | 2.2 A refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1AOL
 
 | | FRIEND MURINE LEUKEMIA VIRUS RECEPTOR-BINDING DOMAIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GP70, ZINC ION | | Authors: | Fass, D, Davey, R.A, Hamson, C.A, Kim, P.S, Cunningham, J.M, Berger, J.M. | | Deposit date: | 1997-07-08 | | Release date: | 1997-10-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 angstrom resolution.
Science, 277, 1997
|
|
1KEY
 
 | | Crystal Structure of Mouse Testis/Brain RNA-binding Protein (TB-RBP) | | Descriptor: | translin | | Authors: | Pascal, J.M, Hart, P.J, Hecht, N.B, Robertus, J.D. | | Deposit date: | 2001-11-19 | | Release date: | 2002-07-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of TB-RBP, a Novel RNA-binding and Regulating Protein
J.Mol.Biol., 319, 2002
|
|
1BYL
 
 | |
8RFM
 
 | | Human NOQ1 enzyme in complex with NADH by serial crystallography | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
8RFN
 
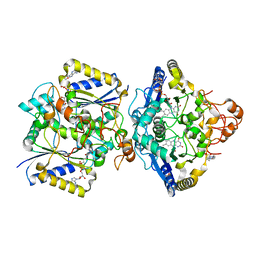 | | Human NOQ1 enzyme in its holo form by serial crystallography | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Martin-Garcia, J.M, Grieco, A, Medina, M, Boneta, S, Pey, A.L. | | Deposit date: | 2023-12-13 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural dynamics and functional cooperativity of human NQO1 by ambient temperature serial crystallography and simulations.
Protein Sci., 33, 2024
|
|
1LAX
 
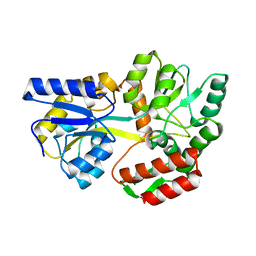 | | CRYSTAL STRUCTURE OF MALE31, A DEFECTIVE FOLDING MUTANT OF MALTOSE-BINDING PROTEIN | | Descriptor: | MALTOSE-BINDING PROTEIN MUTANT MALE31, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Saul, F.A, Mourez, M, Vulliez-le Normand, B, Sassoon, N, Bentley, G.A, Betton, J.M. | | Deposit date: | 2002-03-29 | | Release date: | 2003-03-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of a defective folding protein
PROTEIN SCI., 12, 2003
|
|
1IAM
 
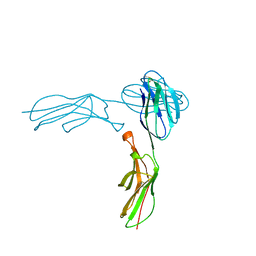 | | STRUCTURE OF THE TWO AMINO-TERMINAL DOMAINS OF HUMAN INTERCELLULAR ADHESION MOLECULE-1, ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERCELLULAR ADHESION MOLECULE-1 | | Authors: | Bella, J, Kolatkar, P.R, Marlor, C, Greve, J.M, Rossmann, M.G. | | Deposit date: | 1998-02-22 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A94
 
 | | STRUCTURAL BASIS FOR SPECIFICITY OF RETROVIRAL PROTEASES | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE | | Authors: | Wu, J, Adomat, J.M, Ridky, T.W, Louis, J.M, Leis, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 1998-04-16 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for specificity of retroviral proteases.
Biochemistry, 37, 1998
|
|
6QM2
 
 | | NlaIV restriction endonuclease | | Descriptor: | POTASSIUM ION, SODIUM ION, Type-2 restriction enzyme NlaIV | | Authors: | Czapinska, H, Siwek, W, Szczepanowski, R.H, Bujnicki, J.M, Bochtler, M, Skowronek, K. | | Deposit date: | 2019-02-01 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure and Directed Evolution of Specificity of NlaIV Restriction Endonuclease.
J.Mol.Biol., 431, 2019
|
|
1K38
 
 | | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2 | | Descriptor: | Beta-lactamase OXA-2, FORMIC ACID | | Authors: | Kerff, F, Fonze, E, Bouillenne, F, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-2
To be Published
|
|
6D43
 
 | |
