8FH1
 
 | |
8FH0
 
 | |
8FGY
 
 | |
4U1T
 
 | |
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
4N5Y
 
 | |
4N5Z
 
 | | Crystal structure of aerosol transmissible influenza H5 hemagglutinin mutant (N158D, N224K, Q226L and T318I) from the influenza virus A/Viet Nam/1203/2004 (H5N1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Zhu, X, Wilson, I.A. | | Deposit date: | 2013-10-10 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9537 Å) | | Cite: | Hemagglutinin Receptor Specificity and Structural Analyses of Respiratory Droplet-Transmissible H5N1 Viruses.
J.Virol., 88, 2014
|
|
7WDF
 
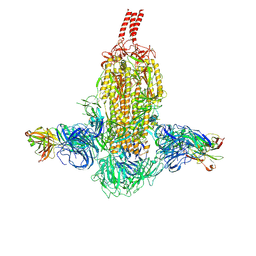 | | SARS-CoV-2 Beta spike in complex with two S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD0
 
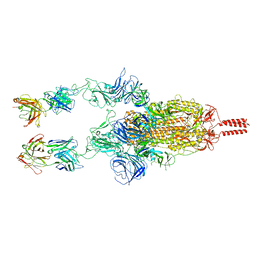 | | SARS-CoV-2 Beta spike in complex with two S5D2 Fabs | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WCR
 
 | |
7WD9
 
 | | SARS-CoV-2 Beta spike in complex with three S3H3 Fabs | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD7
 
 | |
7WCZ
 
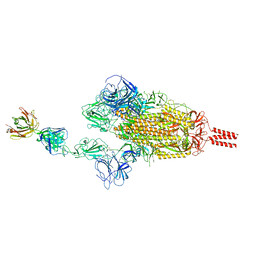 | | SARS-CoV-2 Beta spike in complex with one S5D2 Fab | | Descriptor: | Heavy chain of S5D2 Fab, Light chain of S5D2 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-20 | | Release date: | 2022-02-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
7WD8
 
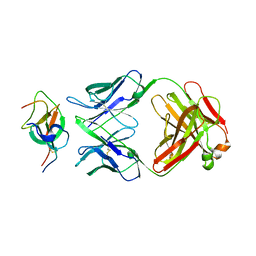 | | SARS-CoV-2 Beta spike SD1 in complex with S3H3 Fab | | Descriptor: | Heavy chain of S3H3 Fab, Light chain of S3H3 Fab, Spike glycoprotein | | Authors: | Wang, Y.F, Cong, Y. | | Deposit date: | 2021-12-21 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping cross-variant neutralizing sites on the SARS-CoV-2 spike protein.
Emerg Microbes Infect, 11, 2022
|
|
5TBM
 
 | | Crystal structure of PT2385 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-{[(1S)-2,2-difluoro-1-hydroxy-7-(methylsulfonyl)-2,3-dihydro-1H-inden-4-yl]oxy}-5-fluorobenzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2016-09-12 | | Release date: | 2016-09-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Small-Molecule Antagonist of HIF2 alpha Is Efficacious in Preclinical Models of Renal Cell Carcinoma.
Cancer Res., 76, 2016
|
|
3DY6
 
 | | PPARdelta complexed with an anthranilic acid partial agonist | | Descriptor: | 2-({[3-(3,4-dihydroisoquinolin-2(1H)-ylsulfonyl)phenyl]carbonyl}amino)benzoic acid, IODIDE ION, Peroxisome proliferator-activated receptor delta | | Authors: | Lambert, M.L, Xu, R, Shearer, B.G, Wilson, T.M. | | Deposit date: | 2008-07-25 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a novel class of PPARdelta partial agonists
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7YUA
 
 | |
7YV0
 
 | |
8WIU
 
 | | Bromodomain and Extra-terminal Domain (BET) BRD4 | | Descriptor: | 7-[5-[1-(cyclopropylmethyl)-3,5-dimethyl-pyrazol-4-yl]pyridin-3-yl]-1~{H}-imidazo[4,5-b]pyridine, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Zhiyan, D, Xiong, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of a brain-permeable bromodomain and extra terminal domain (BET) inhibitor with selectivity for BD1 for the treatment of multiple sclerosis.
Eur.J.Med.Chem., 265, 2023
|
|
3CCB
 
 | | Crystal Structure of Human DPP4 in complex with a benzimidazole derivative | | Descriptor: | 1-biphenyl-2-ylmethanamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wallace, M.B, Skene, R.J. | | Deposit date: | 2008-02-25 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure-based design and synthesis of benzimidazole derivatives as dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2MKZ
 
 | |
3OW3
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OW4
 
 | | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors | | Descriptor: | (2R)-3-(1H-indol-3-yl)-1-{4-[(5S)-5-methyl-5,7-dihydrothieno[3,4-d]pyrimidin-4-yl]piperazin-1-yl}-1-oxopropan-2-amine, GSK 3 beta peptide, RAC-alpha serine/threonine-protein kinase | | Authors: | Dizon, F, Wu, W, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-09-17 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of dihydrothieno- and dihydrofuropyrimidines as potent pan Akt inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3OCB
 
 | | Akt1 kinase domain with pyrrolopyrimidine inhibitor | | Descriptor: | (2S)-2-(4-chlorobenzyl)-3-oxo-3-[4-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperazin-1-yl]propan-1-amine, GSK 3 beta peptide, v-akt murine thymoma viral oncogene homolog 1 (AKT1) | | Authors: | Morales, T.H, Vigers, G.P.A, Brandhuber, B.J. | | Deposit date: | 2010-08-09 | | Release date: | 2010-09-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of pyrrolopyrimidine inhibitors of Akt.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
