8TDI
 
 | | Structure of P2B11 Glucuronide-3-dehydrogenase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, P2B11 Glucuronide-3-dehydrogenase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-03 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCS
 
 | | Structure of trehalose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, Xylose isomerase-like TIM barrel domain-containing protein, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCT
 
 | | Structure of 3K-GlcH bound Bacteroides thetaiotaomicron 3-Keto-beta-glucopyranoside-1,2-Lyase BT1 | | Descriptor: | 1,5-anhydro-D-ribo-hex-3-ulose, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8TCR
 
 | | Structure of glucose bound Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | COBALT (II) ION, MALONATE ION, Sugar phosphate isomerase, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-07-02 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria.
Nature, 631, 2024
|
|
8VA1
 
 | |
8V31
 
 | |
3J1X
 
 | | A refined model of the prototypical Salmonella typhimurium T3SS basal body reveals the molecular basis for its assembly | | Descriptor: | Protein PrgH | | Authors: | Sgourakis, N.G, Bergeron, J.R.C, Worrall, L.J, Strynadka, N.C.J, Baker, D. | | Deposit date: | 2012-07-10 | | Release date: | 2013-05-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | A Refined Model of the Prototypical Salmonella SPI-1 T3SS Basal Body Reveals the Molecular Basis for Its Assembly.
Plos Pathog., 9, 2013
|
|
4G1I
 
 | |
6CO9
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6COJ
 
 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB E105A COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CB2
 
 | | Crystal structure of Escherichia coli UppP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | Authors: | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
6DWB
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome needle filament | | Descriptor: | Protein PrgI | | Authors: | Hu, J, Hong, C, Worrall, L.J, Vuckovic, M, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DV3
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6CON
 
 | | Crystal structure of Mycobacterium tuberculosis IpdAB | | Descriptor: | CoA-transferase subunit alpha, CoA-transferase subunit beta | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6CO6
 
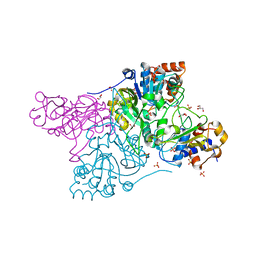 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB | | Descriptor: | GLYCEROL, Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DV6
 
 | | Structure of the Salmonella SPI-1 type III secretion injectisome secretin InvG (residues 176-end) in the open gate state | | Descriptor: | Protein InvG | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
6DUZ
 
 | | Structure of the periplasmic domains of PrgH and PrgK from the assembled Salmonella type III secretion injectisome needle complex | | Descriptor: | Lipoprotein PrgK, Protein PrgH | | Authors: | Hu, J, Worrall, L.J, Vuckovic, M, Atkinson, C.E, Strynadka, N.C.J. | | Deposit date: | 2018-06-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM analysis of the T3S injectisome reveals the structure of the needle and open secretin.
Nat Commun, 9, 2018
|
|
5WC3
 
 | | SpoIIIAG | | Descriptor: | SpoIIIAG, Stage III sporulation engulfment assemblyprotein | | Authors: | Zeytuni, N, Hong, C, Worrall, L.J, Huang, R.K, Yu, Z, Strynadka, N.C.J. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7JP1
 
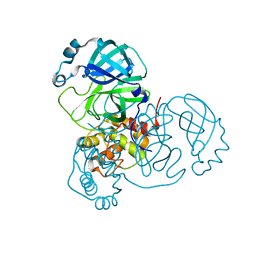 | |
7JOY
 
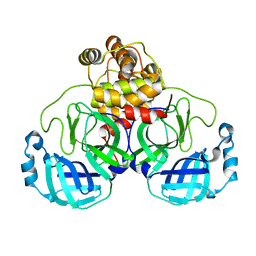 | |
7KHP
 
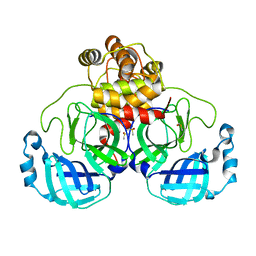 | | Acyl-enzyme intermediate structure of SARS-CoV-2 Mpro in complex with its C-terminal autoprocessing sequence. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Lee, J, Worrall, L.J, Paetzel, M, Strynadka, N.C.J. | | Deposit date: | 2020-10-21 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic structure of wild-type SARS-CoV-2 main protease acyl-enzyme intermediate with physiological C-terminal autoprocessing site.
Nat Commun, 11, 2020
|
|
3T07
 
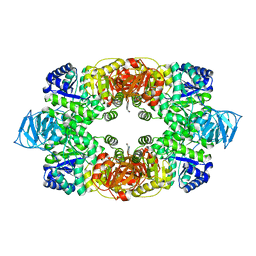 | | Crystal structure of S. aureus Pyruvate Kinase in complex with a naturally occurring bis-indole alkaloid | | Descriptor: | (3S,5R)-3,5-bis(6-bromo-1H-indol-3-yl)piperazin-2-one, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-19 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Methicillin-resistant Staphylococcus aureus (MRSA) pyruvate kinase as a target for bis-indole alkaloids with antibacterial activities.
J.Biol.Chem., 286, 2011
|
|
3T05
 
 | |
3T0T
 
 | | Crystal structure of S. aureus Pyruvate Kinase | | Descriptor: | N'-[(1E)-1-(1H-benzimidazol-2-yl)ethylidene]-5-bromo-2-hydroxybenzohydrazide, PHOSPHATE ION, Pyruvate kinase | | Authors: | Worrall, L.J, Vuckovic, M, Strynadka, N.C.J. | | Deposit date: | 2011-07-20 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Cheminformatics-driven discovery of selective, nanomolar inhibitors for staphylococcal pyruvate kinase.
Acs Chem.Biol., 7, 2012
|
|
