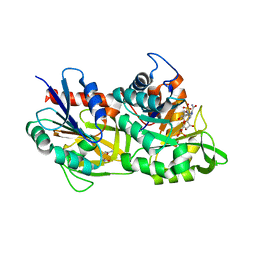
7AC8
 
 | |
1JQQ
 
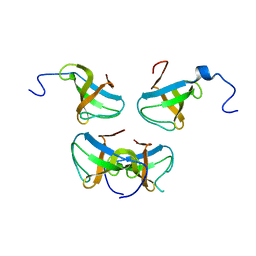 | | Crystal structure of Pex13p(301-386) SH3 domain | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PAS20 | | Authors: | Douangamath, A, Mayans, O, Barnett, P, Distel, B, Wilmanns, M. | | Deposit date: | 2001-08-08 | | Release date: | 2002-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Topography for Independent Binding of alpha-Helical and PPII-Helical Ligands to a Peroxisomal SH3 Domain
Mol.Cell, 10, 2002
|
|
1TKI
 
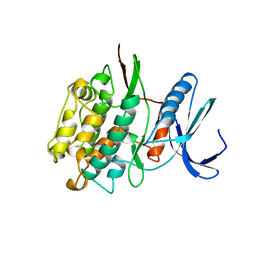 | |
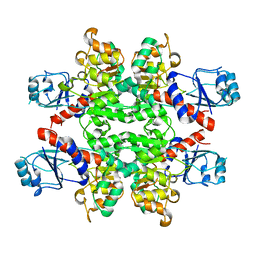
2YQ4
 
 | |
2Y9M
 
 | | Pex4p-Pex22p structure | | Descriptor: | 1,2-ETHANEDIOL, PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-15 | | Release date: | 2011-10-26 | | Last modified: | 2017-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Ubiquitin-Conjugating Enzyme/ Co-Activator Interactions from the Structure of the Pex4P:Pex22P Complex.
Embo J., 31, 2011
|
|
2Y9P
 
 | | Pex4p-Pex22p mutant II structure | | Descriptor: | PEROXISOME ASSEMBLY PROTEIN 22, UBIQUITIN-CONJUGATING ENZYME E2-21 KDA | | Authors: | Williams, C, van den Berg, M, Panjikar, S, Distel, B, Wilmanns, M. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Pex4P-Pex22P Structure
To be Published
|
|
2YQ5
 
 | | Crystal Structure of D-isomer specific 2-hydroxyacid dehydrogenase from Lactobacillus delbrueckii ssp. bulgaricus: NAD complexed form | | Descriptor: | D-ISOMER SPECIFIC 2-HYDROXYACID DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Holton, S.J, Anandhakrishnan, M, Geerlof, A, Wilmanns, M. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural Characterization of D-Isomer Specific 2-Hydroxyacid Dehydrogenase from Lactobacillus Delbrueckii Ssp. Bulgaricus
J.Struct.Biol., 181, 2013
|
|
3ZR4
 
 | | STRUCTURAL EVIDENCE FOR AMMONIA TUNNELING ACROSS THE (BETA-ALPHA)8 BARREL OF THE IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE BIENZYME COMPLEX | | Descriptor: | GLUTAMINE, GLYCEROL, IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISF, ... | | Authors: | Vega, M.C, Kuper, J, Haeger, M.C, Mohrlueder, J, Marquardt, S, Sterner, R, Wilmanns, M. | | Deposit date: | 2011-06-13 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Catalysis Uncoupling in a Glutamine Amidotransferase Bienzyme by Unblocking the Glutaminase Active Site.
Chem.Biol., 19, 2012
|
|
1ABO
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF THE ABL TYROSINE KINASE SH3 DOMAIN WITH 3BP-1 SYNTHETIC PEPTIDE | | Descriptor: | 3BP-1 SYNTHETIC PEPTIDE, 10 RESIDUES, ABL TYROSINE KINASE, ... | | Authors: | Musacchio, A, Wilmanns, M, Saraste, M. | | Deposit date: | 1995-05-19 | | Release date: | 1995-10-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
1ABQ
 
 | |
1M4J
 
 | | CRYSTAL STRUCTURE OF THE N-TERMINAL ADF-H DOMAIN OF MOUSE TWINFILIN ISOFORM-1 | | Descriptor: | A6 gene product | | Authors: | Paavilainen, V.O, Merckel, M.C, Falck, S, Ojala, P.J, Pohl, E, Wilmanns, M, Lappalainen, P. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Conservation Between the Actin Monomer-binding Sites of Twinfilin and Actin-depolymerizing Factor (ADF)/Cofilin
J.Biol.Chem., 277, 2002
|
|
1OOT
 
 | |
3FAV
 
 | | Structure of the CFP10-ESAT6 complex from Mycobacterium tuberculosis | | Descriptor: | 6 kDa early secretory antigenic target, ESAT-6-like protein esxB, IMIDAZOLE, ... | | Authors: | Poulsen, C, Holton, S.J, Wilmanns, M, Song, Y.H. | | Deposit date: | 2008-11-18 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
1PWT
 
 | | THERMODYNAMIC ANALYSIS OF ALPHA-SPECTRIN SH3 AND TWO OF ITS CIRCULAR PERMUTANTS WITH DIFFERENT LOOP LENGTHS: DISCERNING THE REASONS FOR RAPID FOLDING IN PROTEINS | | Descriptor: | ALPHA SPECTRIN | | Authors: | Martinez, J.C, Viguera, A.R, Berisio, R, Wilmanns, M, Mateo, P.L, Filmonov, V.V, Serrano, L. | | Deposit date: | 1998-10-06 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Thermodynamic analysis of alpha-spectrin SH3 and two of its circular permutants with different loop lengths: discerning the reasons for rapid folding in proteins.
Biochemistry, 38, 1999
|
|
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGH
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset I) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
3GWK
 
 | | Structure of the homodimeric WXG-100 family protein from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein SAG1039, SULFATE ION | | Authors: | Poulsen, C, Gries, F, Wilmanns, M, Song, Y.H. | | Deposit date: | 2009-04-01 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
4HGJ
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
1QDL
 
 | | THE CRYSTAL STRUCTURE OF ANTHRANILATE SYNTHASE FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | PROTEIN (ANTHRANILATE SYNTHASE (TRPE-SUBUNIT)), PROTEIN (ANTHRANILATE SYNTHASE (TRPG-SUBUNIT)) | | Authors: | Knoechel, T, Ivens, A, Hester, G, Gonzalez, A, Bauerle, R, Wilmanns, M, Kirschner, K, Jansonius, J.N. | | Deposit date: | 1999-05-20 | | Release date: | 1999-08-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of anthranilate synthase from Sulfolobus solfataricus: functional implications.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
3H7Q
 
 | |
4HGG
 
 | | Crystal structure of P450 BM3 5F5R heme domain variant complexed with styrene | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
4HGI
 
 | | Crystal structure of P450 BM3 5F5 heme domain variant complexed with styrene (dataset II) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450/NADPH-P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
3GWM
 
 | |
2Y88
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE (VARIANT D11N) WITH BOUND PRFAR | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, [(2R,3S,4R,5R)-5-[4-AMINOCARBONYL-5-[[(Z)-[(3R,4R)-3,4-DIHYDROXY-2-OXO-5-PHOSPHONOOXY-PENTYL]IMINOMETHYL]AMINO]IMIDAZOL-1-YL]-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Kuper, J, Due, A.V, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2Y85
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND RCDRP | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, CHLORIDE ION, PHOSPHORIBOSYL ISOMERASE A, ... | | Authors: | Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
