5KSC
 
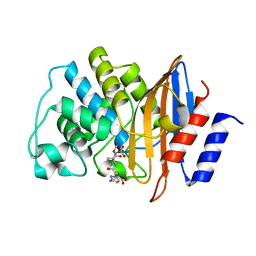 | | E166A/R274N/R276N Toho-1 Beta-lactamase aztreonam acyl-enzyme intermediate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, Beta-lactamase Toho-1 | | Authors: | Vandavasi, V.G, Langan, P.S, Weiss, K, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-07-08 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-09 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
7TFR
 
 | | Room temperature X-ray structure of SARS-CoV-2 main protease (3CL Mpro) in complex with NBH-2 | | Descriptor: | (1R,2S,5S)-N-{(1Z,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-({1-[(2-methylpropane-2-sulfonyl)methyl]cyclohexyl}carbamoyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase | | Authors: | Kovalevsky, A, Kneller, D.W, Coates, L. | | Deposit date: | 2022-01-07 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease
Nat Commun, 13, 2022
|
|
4GD6
 
 | | SHV-1 beta-lactamase in complex with penam sulfone SA1-204 | | Descriptor: | (3R)-N-(2-formylindolizin-3-yl)-4-[(phenylacetyl)oxy]-3-sulfino-D-valine, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE | | Authors: | van den Akker, F, Wei, K. | | Deposit date: | 2012-07-31 | | Release date: | 2013-07-31 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structures of SHV-1 beta-lactamase with penem and penam sulfone inhibitors that form cyclic intermediates stabilized by carbonyl conjugation
Plos One, 7, 2012
|
|
6MO3
 
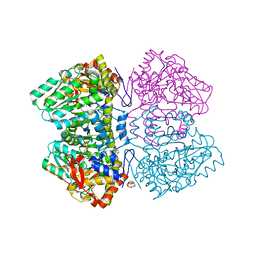 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-serine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-10-04 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MLS
 
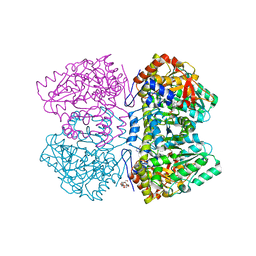 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from L-tyrosine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-09-27 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MPD
 
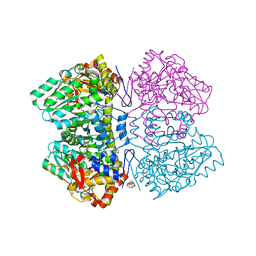 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from 3-F-L-tyrosine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, 3-FLUOROTYROSINE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-10-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MME
 
 | | Citrobacter freundii tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-09-30 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6MQQ
 
 | | Citrobacter freundii F448A mutant tyrosine phenol-lyase complexed with 4-hydroxypyridine and aminoacrylate from S-ethyl-L-cysteine | | Descriptor: | 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, POTASSIUM ION, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2018-10-10 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
6NV8
 
 | | Perdeuterated tyrosine phenol-lyase from Citrobacter freundii complexed with an aminoacrylate intermediate formed from S-ethyl-L-cysteine and 4-hydroxypyridine | | Descriptor: | 2-AMINO-ACRYLIC ACID, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2019-02-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pressure and Temperature Effects on the Formation of Aminoacrylate Intermediates of Tyrosine Phenol-lyase Demonstrate Reaction Dynamics
Acs Catalysis, 10, 2020
|
|
4HK8
 
 | | Crystal Structures of Mutant Endo- -1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | CITRIC ACID, Endo-1,4-beta-xylanase 2, GLYCEROL, ... | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.151 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HK9
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKL
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with substrate (1.15 A) and Products (1.6 A) | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKO
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II (E177Q) in the apo form | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Langan, P, Wan, Q, Coates, L, Kovalevsky, A. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4HKW
 
 | | Crystal Structures of Mutant Endo-beta-1,4-xylanase II Complexed with Substrate and Products | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Endo-1,4-beta-xylanase 2, ... | | Authors: | Kovalevsky, A.Y, Wan, Q, Langan, P, Coates, L. | | Deposit date: | 2012-10-15 | | Release date: | 2014-01-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray crystallographic studies of family 11 xylanase Michaelis and product complexes: implications for the catalytic mechanism.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C9T
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 2.0 A RESOLUTION, SelenoMet derivative | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4C9S
 
 | | BACTERIAL CHALCONE ISOMERASE IN open CONFORMATION FROM EUBACTERIUM RAMULUS AT 1.8 A RESOLUTION | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2013-10-03 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4D06
 
 | | Bacterial chalcone isomerase complexed with naringenin | | Descriptor: | (2E)-3-(4-hydroxyphenyl)-1-(2,4,6-trihydroxyphenyl)prop-2-en-1-one, CHALCONE ISOMERASE, CHLORIDE ION, ... | | Authors: | Thomsen, M, Palm, G.J, Hinrichs, W. | | Deposit date: | 2014-04-24 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of the Evolutionarily Unique Bacterial Chalcone Isomerase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8B7Z
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Palm, G.J, Hinrichs, W. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7R
 
 | | Bacterial chalcone isomerase with taxifolin chalcone | | Descriptor: | (Z)-3-[3,4-bis(oxidanyl)phenyl]-2-oxidanyl-1-[2,4,6-tris(oxidanyl)phenyl]prop-2-en-1-one, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
8B7U
 
 | | Bacterial chalcone isomerase H33A with taxifolin | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, CHLORIDE ION, Chalcone isomerase, ... | | Authors: | Hinrichs, W, Palm, G.J. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus .
Molecules, 27, 2022
|
|
5ZKZ
 
 | |
5ZO0
 
 | | Neutron structure of xylanase at pD5.4 | | Descriptor: | Endo-1,4-beta-xylanase 2 | | Authors: | Wan, Q, Li, Z.H. | | Deposit date: | 2018-04-12 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | NEUTRON DIFFRACTION (1.648 Å) | | Cite: | Neutron structure of xylanase at pD5.4
To be published
|
|
5ZIW
 
 | |
5ZII
 
 | |
6K9R
 
 | | Crystal Structure Analysis of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | Endo-1,4-beta-xylanase 2, IODIDE ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
