4E91
 
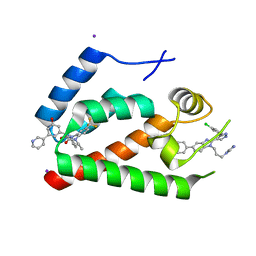 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BD3 | | Descriptor: | (3S)-1-ethyl-3-[3-hydroxy-5-(pyridin-3-yl)phenyl]-5-phenyl-7-(trifluoromethyl)-1H-1,5-benzodiazepine-2,4(3H,5H)-dione, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein, ... | | Authors: | Lemke, C.T. | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
4E92
 
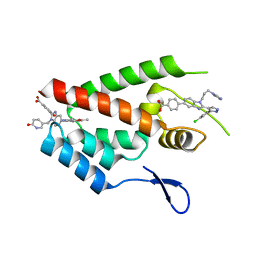 | | Crystal Structure of the N-Terminal Domain of HIV-1 Capsid in Complex With Inhibitor BM4 | | Descriptor: | 3-{5-[3-ethyl-5-(5-methylfuran-2-yl)-1H-pyrazol-1-yl]-1-[(6-oxo-1,6-dihydropyridin-3-yl)methyl]-1H-benzimidazol-2-yl}-4-hydroxybenzoic acid, 4-{2-[5-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid, Gag protein | | Authors: | Lemke, C.T. | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Distinct Effects of Two HIV-1 Capsid Assembly Inhibitor Families That Bind the Same Site within the N-Terminal Domain of the Viral CA Protein.
J.Virol., 86, 2012
|
|
3GV2
 
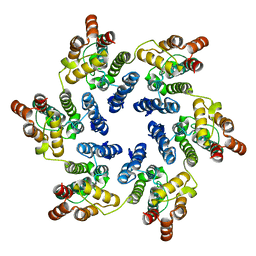 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24,Carbon dioxide-concentrating mechanism protein CcmK homolog 4 | | Authors: | Kelly, B.N. | | Deposit date: | 2009-03-30 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
6TZ9
 
 | |
6TZ4
 
 | |
6TZA
 
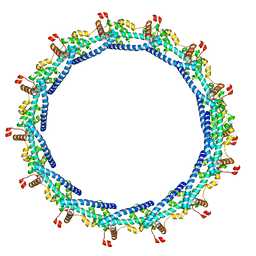 | |
3H4E
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-19 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
6TZ5
 
 | |
6V2F
 
 | | Crystal structure of the HIV capsid hexamer bound to the small molecule long-acting inhibitor, GS-6207 | | Descriptor: | HIV-1 capsid, N-[(1S)-1-(3-{4-chloro-3-[(methylsulfonyl)amino]-1-(2,2,2-trifluoroethyl)-1H-indazol-7-yl}-6-[3-methyl-3-(methylsulfonyl)but-1-yn-1-yl]pyridin-2-yl)-2-(3,5-difluorophenyl)ethyl]-2-[(3bS,4aR)-5,5-difluoro-3-(trifluoromethyl)-3b,4,4a,5-tetrahydro-1H-cyclopropa[3,4]cyclopenta[1,2-c]pyrazol-1-yl]acetamide | | Authors: | Appleby, T.C, Link, J.O, Yant, S.R, Villasenor, A.G, Somoza, J.R, Hu, E.Y, Schroeder, S.D, Cihlar, T. | | Deposit date: | 2019-11-22 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Clinical targeting of HIV capsid protein with a long-acting small molecule.
Nature, 584, 2020
|
|
1A6S
 
 | | M-DOMAIN FROM GAG POLYPROTEIN OF ROUS SARCOMA VIRUS, NMR, 20 STRUCTURES | | Descriptor: | GAG POLYPROTEIN | | Authors: | Mcdonnell, J.M, Fushman, D, Cahill, S.M, Zhou, W, Wolven, A, Wilson, C.B, Nelle, T.D, Resh, M.D, Wills, J, Cowburn, D. | | Deposit date: | 1998-03-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the bioactive retroviral M domain from Rous sarcoma virus
J.Mol.Biol., 279, 1998
|
|
3H47
 
 | | X-ray Structure of Hexameric HIV-1 CA | | Descriptor: | Capsid protein p24 | | Authors: | Pornillos, O. | | Deposit date: | 2009-04-18 | | Release date: | 2009-06-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structures of the hexameric building block of the HIV capsid.
Cell(Cambridge,Mass.), 137, 2009
|
|
4LCB
 
 | | Structure of Vps4 homolog from Acidianus hospitalis | | Descriptor: | CHLORIDE ION, Cell division protein CdvC, Vps4 | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-21 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LGM
 
 | | Crystal Structure of Sulfolobus solfataricus Vps4 | | Descriptor: | CHLORIDE ION, Vps4 AAA ATPase | | Authors: | Han, H, Hill, C.P, Whitby, F.G, Monroe, N. | | Deposit date: | 2013-06-28 | | Release date: | 2013-11-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | The Oligomeric State of the Active Vps4 AAA ATPase.
J.Mol.Biol., 426, 2014
|
|
4LTB
 
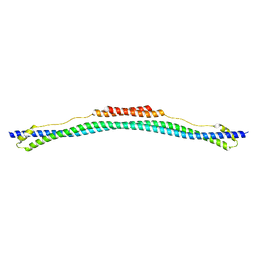 | | Coiled-coil domain of TRIM25 | | Descriptor: | Tripartite motif-containing 25 variant | | Authors: | Pornillos, O, Sanchez, J.G, Okreglicka, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2OEX
 
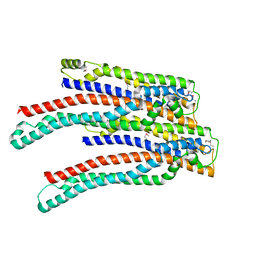 | | Structure of ALIX/AIP1 V Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2OEW
 
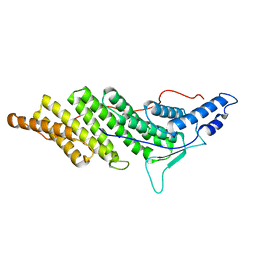 | | Structure of ALIX/AIP1 Bro1 Domain | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2OEV
 
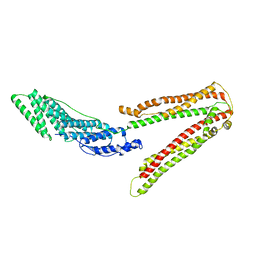 | | Crystal structure of ALIX/AIP1 | | Descriptor: | Programmed cell death 6-interacting protein | | Authors: | Fisher, R.D, Zhai, Q, Robinson, H, Hill, C.P. | | Deposit date: | 2007-01-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Biochemical Studies of ALIX/AIP1 and Its Role in Retrovirus Budding
Cell(Cambridge,Mass.), 128, 2007
|
|
2GOL
 
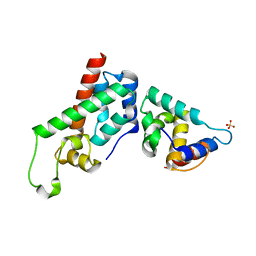 | | Xray Structure of Gag278 | | Descriptor: | Capsid protein p24 (CA), Matrix protein p17 (MA), SULFATE ION | | Authors: | Kelly, B.N. | | Deposit date: | 2006-04-13 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for Viral Capsid Assembly from Crystal Structures of HIV-1 Gag 1-278 and CAN 133-278.
Biochemistry, 45, 2006
|
|
2PWO
 
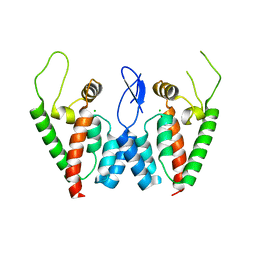 | | Crystal Structure of HIV-1 CA146 A92E Psuedo Cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol) | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2PWM
 
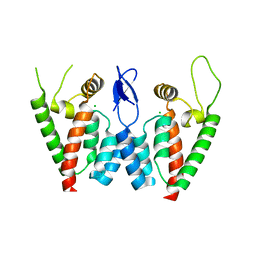 | | Crystal Structure of HIV-1 CA146 A92E real cell | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2PXR
 
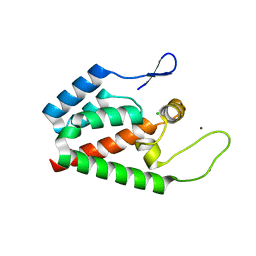 | | Crystal Structure of HIV-1 CA146 in the Presence of CAP-1 | | Descriptor: | CHLORIDE ION, Gag-Pol polyprotein (Pr160Gag-Pol), ZINC ION | | Authors: | Kelly, B.N. | | Deposit date: | 2007-05-14 | | Release date: | 2007-09-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein.
J.Mol.Biol., 373, 2007
|
|
2GON
 
 | | Xray Structure of Gag133-278 | | Descriptor: | CITRATE ANION, Capsid protein p24 (CA) | | Authors: | Kelly, B.N. | | Deposit date: | 2006-04-13 | | Release date: | 2006-09-26 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Implications for Viral Capsid Assembly from Crystal Structures of HIV-1 Gag 1-278 and CAN 133-278.
Biochemistry, 45, 2006
|
|
2R03
 
 | |
2R05
 
 | |
1AWR
 
 | | CYPA COMPLEXED WITH HAGPIA | | Descriptor: | CYCLOPHILIN A, PEPTIDE FROM THE HIV-1 CAPSID PROTEIN | | Authors: | Vajdos, F.F. | | Deposit date: | 1997-10-04 | | Release date: | 1998-03-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structure of cyclophilin A complexed with a binding site peptide from the HIV-1 capsid protein.
Protein Sci., 6, 1997
|
|
