2ZNT
 
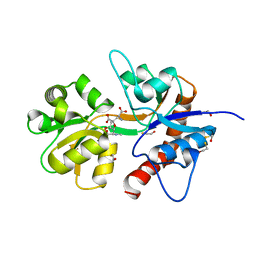 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with a novel selective agonist, dysiherbaine | | Descriptor: | (2R,3aR,6S,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6-hydroxy-7-(methylamino)hexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, BETA-MERCAPTOETHANOL, Glutamate receptor, ... | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
8H3M
 
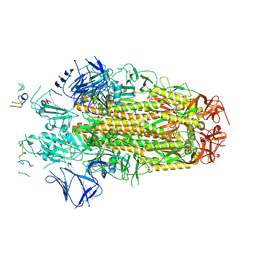 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
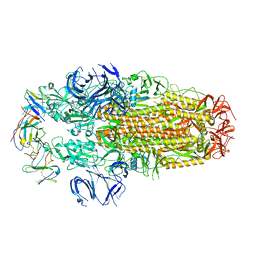 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
2ZNS
 
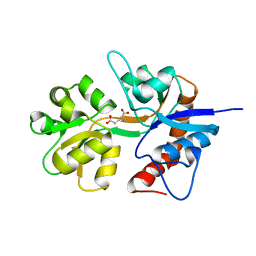 | | Crystal structure of the ligand-binding core of the human ionotropic glutamate receptor, GluR5, in complex with glutamate | | Descriptor: | GLUTAMIC ACID, Glutamate receptor, ionotropic kainate 1 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2008-05-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
5GQC
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, SODIUM ION | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQG
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-galactopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
5GQF
 
 | | Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex | | Descriptor: | CALCIUM ION, Lacto-N-biosidase, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yamada, C, Arakawa, T, Katayama, T, Fushinobu, S. | | Deposit date: | 2016-08-07 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum
Cell Chem Biol, 24, 2017
|
|
3EY2
 
 | |
3EY3
 
 | |
5WZN
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - GalNAc complex | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Alpha-N-acetylgalactosaminidase, CALCIUM ION, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
2Q1R
 
 | | Crystal Structure Analysis of the RNA Dodecamer CGCGAAUUAGCG, with a G-A mismatch. | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*AP*GP*CP*G)-3') | | Authors: | Li, F, Pallan, P.S. | | Deposit date: | 2007-05-25 | | Release date: | 2007-10-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Crystal structure, stability and in vitro RNAi activity of oligoribonucleotides containing the ribo-difluorotoluyl nucleotide: insights into substrate requirements by the human RISC Ago2 enzyme.
Nucleic Acids Res., 35, 2007
|
|
3QXM
 
 | | Crystal Structure of Human GluK2 Ligand-Binding Core in Complex with Novel Marine-Derived Toxins, Neodysiherbaine A | | Descriptor: | (2R,3aR,6R,7R,7aR)-2-[(2S)-2-amino-2-carboxyethyl]-6,7-dihydroxyhexahydro-2H-furo[3,2-b]pyran-2-carboxylic acid, Glutamate receptor ionotropic, kainate 2 | | Authors: | Unno, M, Sasaki, M, Ikeda-Saito, M. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Binding and Selectivity of the Marine Toxin Neodysiherbaine A and Its Synthetic Analogues to GluK1 and GluK2 Kainate Receptors.
J.Mol.Biol., 413, 2011
|
|
2Q1O
 
 | |
3EY1
 
 | |
5WZP
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - ligand free | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZQ
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - quadruple mutant | | Descriptor: | Alpha-N-acetylgalactosaminidase, GLYCEROL, ZINC ION | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5WZR
 
 | | Alpha-N-acetylgalactosaminidase NagBb from Bifidobacterium bifidum - Gal-NHAc-DNJ complex | | Descriptor: | Alpha-N-acetylgalactosaminidase, CALCIUM ION, N-[(3S,4R,5S,6R)-4,5-dihydroxy-6-(hydroxymethyl)piperidin-3-yl]acetamide, ... | | Authors: | Sato, M, Arakawa, T, Ashida, H, Fushinobu, S. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | The first crystal structure of a family 129 glycoside hydrolase from a probiotic bacterium reveals critical residues and metal cofactors
J. Biol. Chem., 292, 2017
|
|
5V1L
 
 | | Structure of S-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*UP*(ZTH)P*AP*GP*CP*G)-3'), SPERMINE, STRONTIUM ION | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
5V1K
 
 | | Structure of R-GNA dodecamer | | Descriptor: | RNA (5'-R(*CP*GP*CP*GP*AP*AP*(5BU)P*(8RJ)P*AP*GP*CP*G)-3') | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2017-03-02 | | Release date: | 2017-06-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 2017
|
|
5V2H
 
 | | RNA octamer containing glycol nucleic acid, SgnT | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, RNA (5'-R(*(CBV)P*GP*AP*AP*(ZTH)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2017-03-04 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.080013 Å) | | Cite: | Chirality Dependent Potency Enhancement and Structural Impact of Glycol Nucleic Acid Modification on siRNA.
J. Am. Chem. Soc., 139, 2017
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
468D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
470D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
471D
 
 | | CRYSTAL STRUCTURE AND IMPROVED ANTISENSE PROPERTIES OF 2'-O-(2-METHOXYETHYL)-RNA | | Descriptor: | MAGNESIUM ION, RNA (5'-R(*(C43)P*(G48)P*(C43)P*(G48)P*(A44)P*(A44)P*(U36)P*(U36)P*(C43)P*(G48)P*(C43)P*(G48))-3') | | Authors: | Teplova, M, Minasov, G, Tereshko, V, Inamati, G, Cook, P.D, Egli, M. | | Deposit date: | 1999-04-29 | | Release date: | 1999-05-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and improved antisense properties of 2'-O-(2-methoxyethyl)-RNA.
Nat.Struct.Biol., 6, 1999
|
|
7JJE
 
 | |
