4L1K
 
 | |
3PH3
 
 | | Clostridium thermocellum Ribose-5-Phosphate Isomerase B with d-ribose | | Descriptor: | D-ribose, Ribose-5-phosphate isomerase | | Authors: | Jung, J, Kim, J.K, Yeom, S.J, Ahn, Y.J, Oh, D.K, Kang, L.W. | | Deposit date: | 2010-11-03 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of Clostridium thermocellum ribose-5-phosphate isomerase B reveals properties critical for fast enzyme kinetics.
Appl.Microbiol.Biotechnol., 90, 2011
|
|
3RFC
 
 | |
3R5F
 
 | |
2N0A
 
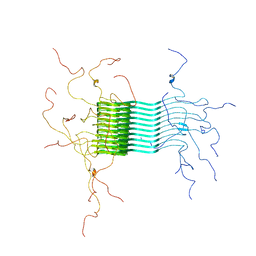 | | Atomic-resolution structure of alpha-synuclein fibrils | | Descriptor: | Alpha-synuclein | | Authors: | Tuttle, M.D, Comellas, G, Nieuwkoop, A.J, Covell, D.J, Berthold, D.A, Kloepper, K.D, Courtney, J.M, Kim, J.K, Schwieters, C.D, Lee, V.M, George, J.M, Rienstra, C.M. | | Deposit date: | 2015-03-04 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Solid-state NMR structure of a pathogenic fibril of full-length human alpha-synuclein.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3HR4
 
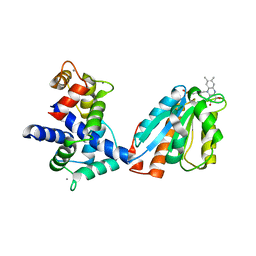 | | Human iNOS Reductase and Calmodulin Complex | | Descriptor: | CALCIUM ION, Calmodulin, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Xia, C, Misra, I, Iyanaki, T, Kim, J.J.K. | | Deposit date: | 2009-06-08 | | Release date: | 2009-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulation of Interdomain Interactions by CaM in Inducible Nitric Oxide Synthase
J.Biol.Chem., 2009
|
|
5WE1
 
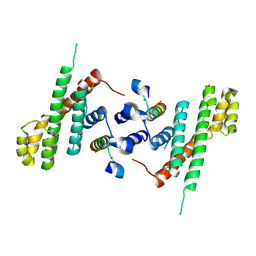 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | Protection of telomeres protein poz1,Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ZINC ION | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5WE0
 
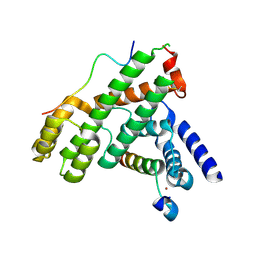 | | Structural Basis for Shelterin Bridge Assembly | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Yu, C, Roskamp, K, Sankaran, B, Huang, L, Komives, E.-A, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
5Y2R
 
 | | 2.5 atm CO2-pressurized human carbonic anhydrase II | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y. | | Deposit date: | 2017-07-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Active-site solvent replenishment observed during human carbonic anhydrase II catalysis.
IUCrJ, 5, 2018
|
|
5Y2S
 
 | | 7.0 atm CO2-pressurized human carbonic anhydrase II | | Descriptor: | CARBON DIOXIDE, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Kim, C.U, Park, S.Y. | | Deposit date: | 2017-07-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Active-site solvent replenishment observed during human carbonic anhydrase II catalysis.
IUCrJ, 5, 2018
|
|
5WE2
 
 | | Structural Basis for Telomere Length Regulation by the Shelterin Bridge | | Descriptor: | DNA-binding protein rap1, Protection of telomeres protein poz1, Protection of telomeres protein tpz1, ... | | Authors: | Kim, J.-K, Liu, J, Hu, X, Sankaran, B, Qiao, F. | | Deposit date: | 2017-07-06 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Shelterin Bridge Assembly.
Mol. Cell, 68, 2017
|
|
6U7V
 
 | | xRRM structure of spPof8 | | Descriptor: | NITRATE ION, Protein pof8 | | Authors: | Kim, J.-K, Hu, X, Yu, C, Jun, H.-I, Liu, J, Sankaran, B, Huang, L, Qiao, F. | | Deposit date: | 2019-09-03 | | Release date: | 2020-09-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Quality-Control Mechanism for Telomerase RNA Folding in the Cell.
Cell Rep, 33, 2020
|
|
8SD8
 
 | | Carbonic anhydrase II radiation damage RT 91-120 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD7
 
 | | Carbonic anhydrase II radiation damage RT 61-90 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD6
 
 | | Carbonic anhydrase II radiation damage RT 31-60 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD9
 
 | | Carbonic anhydrase II radiation damage RT 121-150 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SD1
 
 | | Carbonic anhydrase II radiation damage RT 1-30 | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.C, Mckenna, R. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8SF1
 
 | | Carbonic anhydrase II XFEL radiation damage RT | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Combs, J.E, Mckenna, R. | | Deposit date: | 2023-04-10 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | XFEL structure of carbonic anhydrase II: a comparative study of XFEL, NMR, X-ray and neutron structures.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6JL9
 
 | | Crystal structure of a frog ependymin related protein | | Descriptor: | CALCIUM ION, Ependymin-related 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-04 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JLD
 
 | | Crystal structure of a human ependymin related protein | | Descriptor: | Mammalian ependymin-related protein 1 | | Authors: | Park, S.Y. | | Deposit date: | 2019-03-05 | | Release date: | 2019-07-10 | | Last modified: | 2019-07-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
6JLA
 
 | | Crystal structure of a mouse ependymin related protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Mammalian ependymin-related protein 1 | | Authors: | Park, S. | | Deposit date: | 2019-03-04 | | Release date: | 2020-03-04 | | Last modified: | 2020-09-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of three ependymin-related proteins suggest their function as a hydrophobic molecule binder.
Iucrj, 6, 2019
|
|
3UU0
 
 | | Crystal structure of L-rhamnose isomerase from Bacillus halodurans in complex with Mn | | Descriptor: | L-rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-27 | | Release date: | 2012-02-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UVA
 
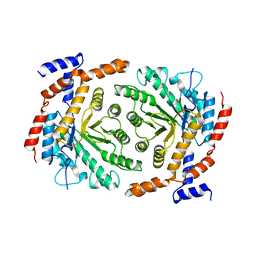 | | Crystal structure of L-rhamnose isomerase mutant W38F from Bacillus halodurans in complex with Mn | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Jeya, M, Kim, J.K, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-11-29 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
3UXI
 
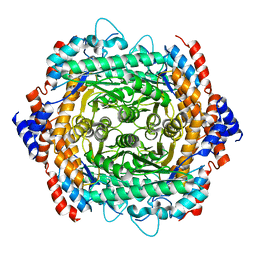 | | Crystal structure of L-rhamnose isomerase W38A mutant from Bacillus halodurans | | Descriptor: | L-Rhamnose isomerase, MANGANESE (II) ION | | Authors: | Doan, T.T.N, Prabhu, P, Kim, J.K, Jeya, M, Kang, L.W, Lee, J.K. | | Deposit date: | 2011-12-05 | | Release date: | 2012-12-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structure-based studies on the metal binding of two-metal-dependent sugar isomerases.
Febs J., 281, 2014
|
|
9BHI
 
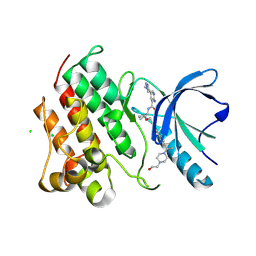 | | Crystal structure of the MerTK kinase domain with SA4488 | | Descriptor: | (5P)-2-amino-5-(1-methyl-1H-pyrazol-4-yl)-N-{(1R,2S)-2-[(4'-{2-[4-(2-oxoethyl)piperazin-1-yl]propan-2-yl}[1,1'-biphenyl]-4-yl)methoxy]cyclopentyl}pyridine-3-carboxamide, CHLORIDE ION, Mer tyrosine kinase domain | | Authors: | Jakob, C.G, Qui, W, Jain, R. | | Deposit date: | 2024-04-20 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of A-910, a Highly Potent and Orally Bioavailable Dual MerTK/Axl-Selective Tyrosine Kinase Inhibitor.
J.Med.Chem., 2024
|
|
