2YXM
 
 | | Crystal structure of I-set domain of human Myosin Binding ProteinC | | Descriptor: | Myosin-binding protein C, slow-type | | Authors: | Kishishita, S, Ohsawa, N, Murayama, K, Chen, L, Liu, Z, Terada, T, Shirouzu, M, Wang, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2007-10-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structure of I-set domain of human Myosin Binding ProteinC
To be Published
|
|
2YV8
 
 | | Crystal structure of N-terminal domain of human galectin-8 | | Descriptor: | Galectin-8 variant | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of N-terminal domain of human galectin-8
To be Published
|
|
2YXS
 
 | | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose | | Descriptor: | Galectin-8 variant, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Kishishita, S, Nishino, A, Murayama, K, Terada, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-27 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Crystal Structure of N-terminal domain of human galectin-8 with D-lactose
To be Published
|
|
2YYO
 
 | | Crystal structure of human SPRY domain | | Descriptor: | SPRY domain-containing protein 3 | | Authors: | Kishishita, S, Uchikubo-Kamo, T, Murayama, K, Terada, T, Chen, L, Fu, Z.Q, Chrzas, J, Shirouzu, M, Wang, B.C, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-30 | | Release date: | 2008-05-06 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human SPRY domain
To be Published
|
|
3VSE
 
 | | Crystal structure of methyltransferase | | Descriptor: | Putative uncharacterized protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kita, S, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2012-04-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of a putative methyltransferase SAV1081 from Staphylococcus aureus
Protein Pept.Lett., 20, 2012
|
|
2DX6
 
 | | Crystal structure of conserved hypothetical protein, TTHA0132 from Thermus thermophilus HB8 | | Descriptor: | ACETATE ION, Hypothetical protein TTHA0132, ISOPROPYL ALCOHOL | | Authors: | Kishishita, S, Murayama, K, Terada, T, Shirouzu, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-24 | | Release date: | 2007-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of conserved hypothetical protein, TTHA0132 from Thermus thermophilus HB8
To be Published
|
|
2CXD
 
 | | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8 | | Descriptor: | conserved hypothetical protein, TTHA0068 | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8
To be Published
|
|
2CWY
 
 | | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA0068 | | Authors: | Kishishita, S, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-12-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of conserved hypothetical protein, TTHA0068 from Thermus thermophilus HB8
To be Published
|
|
2E1D
 
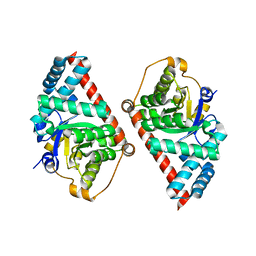 | | Crystal structure of mouse transaldolase | | Descriptor: | SULFITE ION, Transaldolase | | Authors: | Kishishita, S, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-10-25 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mouse transaldolase
To be Published
|
|
2EKK
 
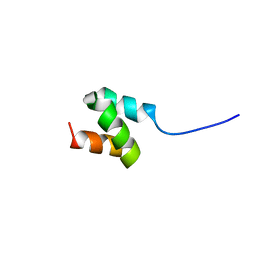 | | Solution structure of RUH-074, a human UBA domain | | Descriptor: | UBA domain from E3 ubiquitin-protein ligase HUWE1 | | Authors: | Kitasaka, S, Ruhul Momen, A.Z.M, Hirota, H, Muto, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RUH-074, a human UBA domain
to be published
|
|
2FEB
 
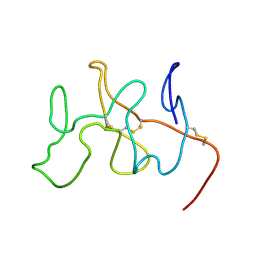 | | NMR Solution Structure, Dynamics and Binding Properties of the Kringle IV Type 8 module of apolipoprotein(a) | | Descriptor: | Apolipoprotein(a) | | Authors: | Chitayat, S, Kanelis, V, Koschinsky, M.L, Smith, S.P. | | Deposit date: | 2005-12-15 | | Release date: | 2006-12-26 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance (NMR) solution structure, dynamics, and binding properties of the kringle IV type 8 module of apolipoprotein(a).
Biochemistry, 46, 2007
|
|
2GML
 
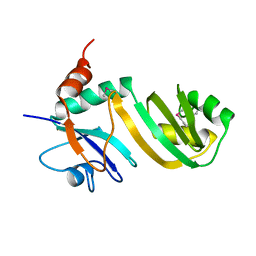 | | Crystal Structure of Catalytic Domain of E.coli RluF | | Descriptor: | Ribosomal large subunit pseudouridine synthase F | | Authors: | Sunita, S, Zhenxing, H, Swaathi, J, Cygler, M, Matte, A, Sivaraman, J. | | Deposit date: | 2006-04-06 | | Release date: | 2006-07-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Domain Organization and Crystal Structure of the Catalytic Domain of E.coli RluF, a Pseudouridine Synthase that Acts on 23S rRNA
J.Mol.Biol., 359, 2006
|
|
2D3W
 
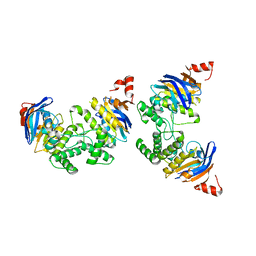 | | Crystal Structure of Escherichia coli SufC, an ATPase compenent of the SUF iron-sulfur cluster assembly machinery | | Descriptor: | Probable ATP-dependent transporter sufC | | Authors: | Kitaoka, S, Wada, K, Hasegawa, Y, Minami, Y, Takahashi, Y, Fukuyama, K. | | Deposit date: | 2005-10-03 | | Release date: | 2006-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Escherichia coli SufC, an ABC-type ATPase component of the SUF iron-sulfur cluster assembly machinery
Febs Lett., 580, 2006
|
|
3WFR
 
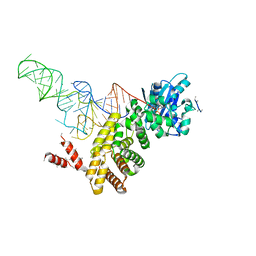 | | tRNA processing enzyme complex 2 | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFS
 
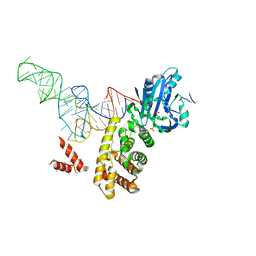 | | tRNA processing enzyme complex 3 | | Descriptor: | Poly A polymerase, RNA (74-MER), SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.311 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFP
 
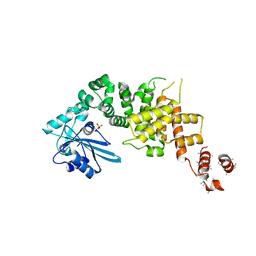 | | tRNA processing enzyme (apo form 2) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFO
 
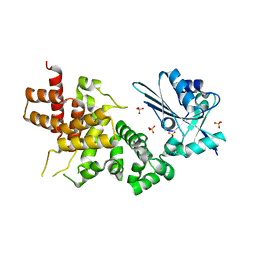 | | tRNA processing enzyme (apo form 1) | | Descriptor: | Poly A polymerase, SULFATE ION | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3WFQ
 
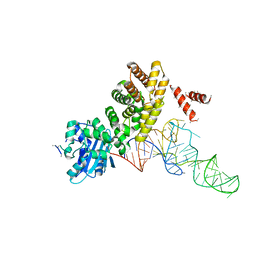 | | tRNA processing enzyme complex 1 | | Descriptor: | Poly A polymerase, RNA (73-MER) | | Authors: | Yamashita, S, Takeshita, D, Tomita, K. | | Deposit date: | 2013-07-23 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.619 Å) | | Cite: | Translocation and rotation of tRNA during template-independent RNA polymerization by tRNA nucleotidyltransferase
Structure, 22, 2014
|
|
3VM8
 
 | |
3VOW
 
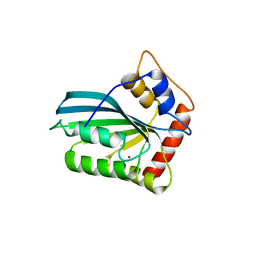 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5Y0P
 
 | |
5Y0S
 
 | |
5Y0R
 
 | |
5Y0N
 
 | |
5Y0Q
 
 | | Crystal structure of BsTmcAL bound with AMPCPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, UPF0348 protein B4417_3650 | | Authors: | Yamashita, S, Tomita, K. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis.
Nat. Chem. Biol., 14, 2018
|
|
