3F8W
 
 | | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase in complex with adenosine | | Descriptor: | ADENOSINE, DIMETHYL SULFOXIDE, Purine-nucleoside phosphorylase, ... | | Authors: | Pereira, H.M, Rezende, M.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-11-13 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1DEA
 
 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1TO4
 
 | | Structure of the cytosolic Cu,Zn SOD from S. mansoni | | Descriptor: | COPPER (II) ION, Superoxide dismutase, ZINC ION | | Authors: | Cardoso, R.M.F, Silva, C.H.T.P, Ulian de Araujo, A.P, Tanaka, T, Tanaka, M, Garratt, R.C. | | Deposit date: | 2004-06-12 | | Release date: | 2004-08-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of the cytosolic Cu,Zn superoxide dismutase from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1TCU
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with phosphate and acetate | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1TD1
 
 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with acetate | | Descriptor: | ACETATE ION, purine-nucleoside phosphorylase | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
1HOR
 
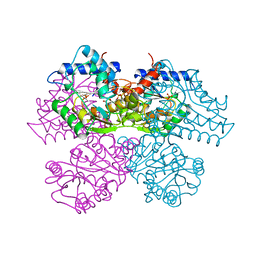 | | STRUCTURE AND CATALYTIC MECHANISM OF GLUCOSAMINE 6-PHOSPHATE DEAMINASE FROM ESCHERICHIA COLI AT 2.1 ANGSTROMS RESOLUTION | | Descriptor: | 2-DEOXY-2-AMINO GLUCITOL-6-PHOSPHATE, GLUCOSAMINE 6-PHOSPHATE DEAMINASE, PHOSPHATE ION | | Authors: | Oliva, G, Fontes, M.R.M, Garratt, R.C, Altamirano, M.M, Calcagno, M.L, Horjales, E. | | Deposit date: | 1995-09-13 | | Release date: | 1996-01-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and catalytic mechanism of glucosamine 6-phosphate deaminase from Escherichia coli at 2.1 A resolution.
Structure, 3, 1995
|
|
1TCV
 
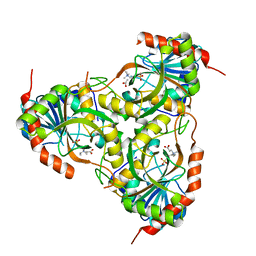 | | Crystal Structure of the Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with Non-detergent Sulfobetaine 195 and acetate | | Descriptor: | ACETATE ION, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, purine-nucleoside phosphorylase | | Authors: | Pereira, H.D, Franco, G.R, Cleasby, A, Garratt, R.C. | | Deposit date: | 2004-05-21 | | Release date: | 2005-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structures for the Potential Drug Target Purine Nucleoside Phosphorylase from Schistosoma mansoni Causal Agent of Schistosomiasis.
J.Mol.Biol., 353, 2005
|
|
4KVA
 
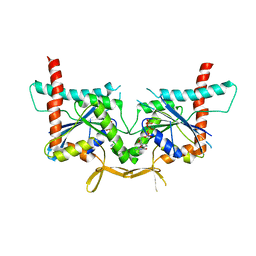 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
4KV9
 
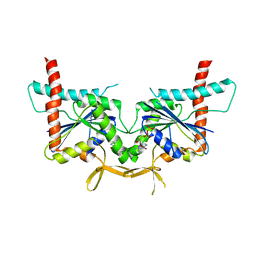 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
1OEP
 
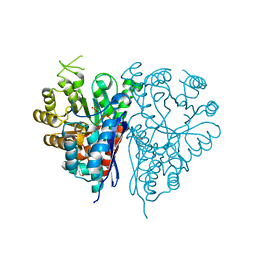 | | Structure of Trypanosoma brucei enolase reveals the inhibitory divalent metal site | | Descriptor: | 1,2-ETHANEDIOL, ENOLASE, SULFATE ION, ... | | Authors: | Da Silva giotto, M.T, Navarro, M.V.A.S, Garratt, R.C, Rigden, D.J. | | Deposit date: | 2003-03-28 | | Release date: | 2003-04-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of Trypanosoma Brucei Enolase: Visualisation of the Inhibitory Metal Binding Site III and Potential as Target for Selective, Irreversible Inhibition
J.Mol.Biol., 331, 2003
|
|
3E9R
 
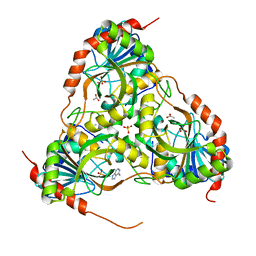 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with adenine | | Descriptor: | ACETATE ION, ADENINE, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3ESF
 
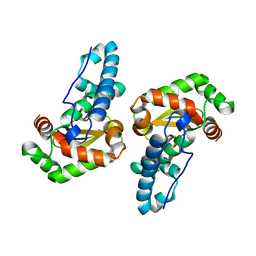 | |
3FNQ
 
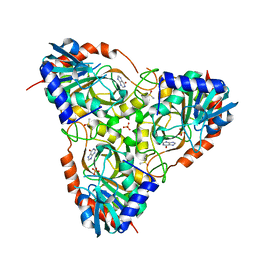 | | Crystal structure of schistosoma purine nucleoside phosphorylase in complex with hypoxanthine | | Descriptor: | DIMETHYL SULFOXIDE, HYPOXANTHINE, Purine-nucleoside phosphorylase, ... | | Authors: | Castilho, M.S, Pereira, H.M, Garratt, R.C, Oliva, G. | | Deposit date: | 2008-12-26 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Adenosine binding to low-molecular-weight purine nucleoside phosphorylase: the structural basis for recognition based on its complex with the enzyme from Schistosoma mansoni.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3E9Z
 
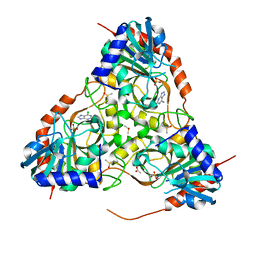 | | Crystal structure of purine nucleoside phosphorylase from Schistosoma mansoni in complex with 6-chloroguanine | | Descriptor: | 6-chloroguanine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pereira, H.M, Rezende, M.M, Oliva, G, Garratt, R.C. | | Deposit date: | 2008-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of Schistosoma mansoni purine nucleoside phosphorylase (SmPNP) in complex with adenine, 8-aminoguanine, 8-azaguanine and 6-chloroguanine.
To be Published
|
|
1CD5
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER | | Descriptor: | PROTEIN (GLUCOSAMINE 6-PHOSPHATE DEAMINASE) | | Authors: | Horjales, E, Altamirano, M.M, Calcagno, M.L, Garratt, R.C, Oliva, G. | | Deposit date: | 1999-03-05 | | Release date: | 2000-03-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The allosteric transition of glucosamine-6-phosphate deaminase: the structure of the T state at 2.3 A resolution.
Structure Fold.Des., 7, 1999
|
|
3SOP
 
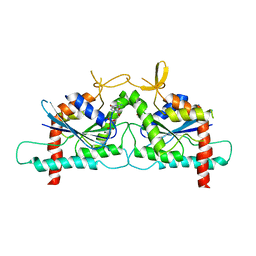 | | Crystal Structure Of Human Septin 3 GTPase Domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Neuronal-specific septin-3 | | Authors: | Marques, I.A, Macedo, J.N.A, Pereira, H.M, Valadares, N.F, Araujo, A.P.U, Garratt, R.C. | | Deposit date: | 2011-06-30 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.885 Å) | | Cite: | The structure and properties of septin 3: a possible missing link in septin filament formation.
Biochem.J., 450, 2013
|
|
3TW4
 
 | | Crystal Structure of Human Septin 7 GTPase Domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin-7 | | Authors: | Serrao, V.H.B, Alessandro, F, Pereira, H.M, Thiemann, O.T, Garratt, R.C. | | Deposit date: | 2011-09-21 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Promiscuous interactions of human septins: The GTP binding domain of SEPT7 forms filaments within the crystal.
Febs Lett., 585, 2011
|
|
3TFB
 
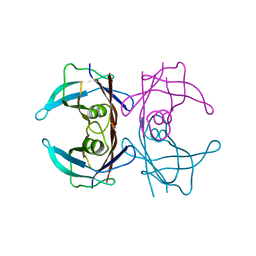 | | Transthyretin natural mutant A25T | | Descriptor: | Transthyretin | | Authors: | Azevedo, E.P.C, Pereira, H.M, Garratt, R.C, Kelly, J.W, Foguel, D, Palhano, F.L. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Dissecting the Structure, Thermodynamic Stability, and Aggregation Properties of the A25T Transthyretin (A25T-TTR) Variant Involved in Leptomeningeal Amyloidosis: Identifying Protein Partners That Co-Aggregate during A25T-TTR Fibrillogenesis in Cerebrospinal Fluid.
Biochemistry, 50, 2011
|
|
3UMP
 
 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with Cesium and ATP | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, CESIUM ION, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Garratt, R.C, Babul, J. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | A Ribokinase Family Conserved Monovalent Cation Binding Site Enhances the MgATP-induced Inhibition in E. coli Phosphofructokinase-2
Biophys.J., 105, 2013
|
|
3UMO
 
 | | Crystal structure of the Phosphofructokinase-2 from Escherichia coli in complex with Potassium | | Descriptor: | 6-phosphofructokinase isozyme 2, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Pereira, H.M, Caniuguir, A, Baez, M, Cabrera, R, Garratt, R.C, Babul, J. | | Deposit date: | 2011-11-14 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | A Ribokinase Family Conserved Monovalent Cation Binding Site Enhances the MgATP-induced Inhibition in E. coli Phosphofructokinase-2
Biophys.J., 105, 2013
|
|
3UL6
 
 | | Saccharum officinarum canecystatin-1 in space group P6422 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Canecystatin-1 | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-12-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
3UL5
 
 | | Saccharum officinarum canecystatin-1 in space group C2221 | | Descriptor: | Canecystatin-1, GLYCEROL, SODIUM ION | | Authors: | Valadares, N.F, Pereira, H.M, Oliveira-Silva, R, Garratt, R.C. | | Deposit date: | 2011-11-10 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystallography and NMR studies of domain-swapped canecystatin-1.
Febs J., 280, 2013
|
|
1S8H
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, first fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8G
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, fatty acid bound form | | Descriptor: | GLYCEROL, LAURIC ACID, Phospholipase A2 homolog, ... | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
1S8I
 
 | | Crystal structure of Lys49-Phospholipase A2 from Agkistrodon contortrix laticinctus, second fatty acid free form | | Descriptor: | Phospholipase A2 homolog, SULFATE ION | | Authors: | Ambrosio, A.L.B, de Souza, D.H.F, Nonato, M.C, Selistre de Araujo, H.S, Ownby, C.L, Garratt, R.C. | | Deposit date: | 2004-02-02 | | Release date: | 2004-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.609 Å) | | Cite: | A Molecular Mechanism for Lys49-Phospholipase A2 Activity Based on Ligand-induced Conformational Change.
J.Biol.Chem., 280, 2005
|
|
