6KYC
 
 | | Structure of the S207A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-17 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
6KYD
 
 | | Structure of the R217A mutant of Clostridium difficile sortase B | | Descriptor: | Putative peptidase C60B, sortase B | | Authors: | Kang, C.Y, Huang, I.H, Wu, T.Y, Chang, J.C, Hsiao, Y.Y, Cheng, C.H, Tsai, W.J, Hsu, K.C, Wang, S.Y. | | Deposit date: | 2019-09-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Functional analysis ofClostridium difficilesortase B reveals key residues for catalytic activity and substrate specificity.
J.Biol.Chem., 295, 2020
|
|
2DF7
 
 | | Crystal structure of infectious bursal disease virus VP2 subviral particle | | Descriptor: | CALCIUM ION, CHLORIDE ION, structural polyprotein VP2 | | Authors: | Ko, T.P, Lee, C.C, Wang, M.Y, Wang, A.H. | | Deposit date: | 2006-02-27 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of infectious bursal disease virus VP2 subviral particle at 2.6A resolution: Implications in virion assembly and immunogenicity.
J.Struct.Biol., 155, 2006
|
|
1WL2
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R90A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL1
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima H74A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL0
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R44A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WL3
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima R91A mutant | | Descriptor: | octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
1WKZ
 
 | | Crystal Structure Of Octaprenyl Pyrophosphate Synthase From Hyperthermophilic Thermotoga Maritima K41A mutant | | Descriptor: | SULFATE ION, octoprenyl-diphosphate synthase | | Authors: | Guo, R.T, Kuo, C.J, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2004-06-18 | | Release date: | 2005-06-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Biochemical and Structural Basis for Octaprenyl Pyrophosphate Synthase
To be Published
|
|
3WUR
 
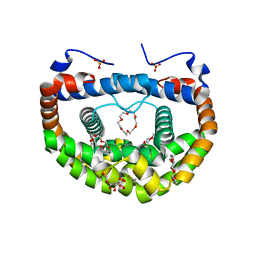 | | Structure of DMP19 Complex with 18-crown-6 | | Descriptor: | 1,2-ETHANEDIOL, 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, L(+)-TARTARIC ACID, ... | | Authors: | Lee, C.C, Wang, H.C, Wang, A.H.J. | | Deposit date: | 2014-05-02 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WH0
 
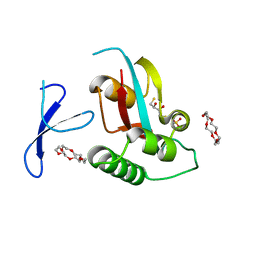 | | Structure of Pin1 Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Lee, C.C, Liu, C.I, Jeng, W.Y, Wang, A.H.J. | | Deposit date: | 2013-08-20 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WHM
 
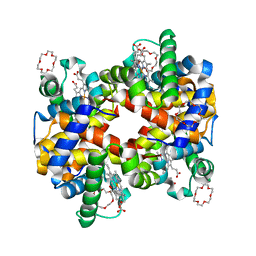 | | Structure of Hemoglobin Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2PGI
 
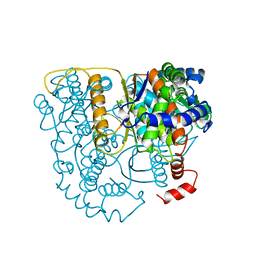 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE-AN ENZYME WITH AUTOCRINE MOTILITY FACTOR ACTIVITY IN TUMOR CELLS | | Descriptor: | PHOSPHOGLUCOSE ISOMERASE | | Authors: | Sun, Y.-J, Chou, C.-C, Chen, W.-S, Meng, M, Hsiao, C.-D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of a multifunctional protein: phosphoglucose isomerase/autocrine motility factor/neuroleukin.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1WD0
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*TP*AP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WD1
 
 | | Crystal structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers | | Descriptor: | 5'-D(*CP*CP*TP*AP*CP*GP*TP*AP*GP*G)-3', DNA-binding proteins 7a/7b/7d | | Authors: | Ko, T.-P, Chu, H.-M, Chen, C.-Y, Chou, C.-C, Wang, A.H.-J. | | Deposit date: | 2004-05-10 | | Release date: | 2004-08-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the hyperthermophilic chromosomal protein Sac7d in complex with DNA decamers.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1E9L
 
 | |
1WVL
 
 | |
1Z1I
 
 | | Crystal structure of native SARS CLpro | | Descriptor: | 3C-like proteinase | | Authors: | Liang, P.H, Wang, A.H. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
1Z1J
 
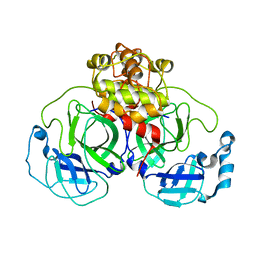 | | Crystal structure of SARS 3CLpro C145A mutant | | Descriptor: | 3C-like proteinase | | Authors: | Hsu, M.F. | | Deposit date: | 2005-03-04 | | Release date: | 2005-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Understanding the maturation process and inhibitor design of SARS-CoV 3CLpro from the crystal structure of C145A in a product-bound form
J.Biol.Chem., 280, 2005
|
|
4QUM
 
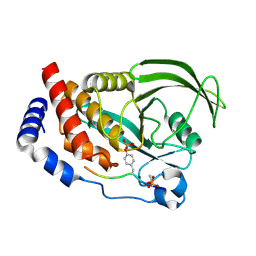 | |
3O98
 
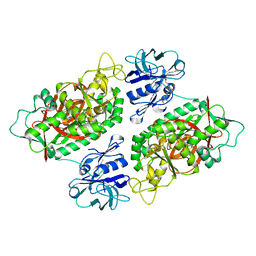 | | Glutathionylspermidine synthetase/amidase C59A complex with ADP and Gsp | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Bifunctional glutathionylspermidine synthetase/amidase, GLUTATHIONYLSPERMIDINE, ... | | Authors: | Pai, C.H, Lin, C.H, Wang, A.H.-J. | | Deposit date: | 2010-08-04 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and mechanism of Escherichia coli glutathionylspermidine amidase belonging to the family of cysteine; histidine-dependent amidohydrolases/peptidases
Protein Sci., 20, 2011
|
|
4RH5
 
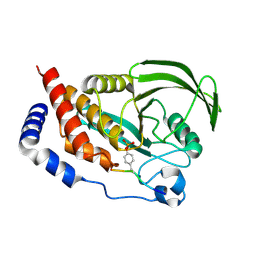 | |
4RH9
 
 | |
4RHG
 
 | |
4RI4
 
 | |
4RI5
 
 | |
