1S6O
 
 | | Solution structure and backbone dynamics of the apo-form of the second metal-binding domain of the Menkes protein ATP7A | | Descriptor: | Copper-transporting ATPase 1 | | Authors: | Banci, L, Bertini, I, Del Conte, R, D'Onofrio, M, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-01-26 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Cu(I) and Apo Forms of the Second Metal-Binding Domain of the Menkes Protein ATP7A.
Biochemistry, 43, 2004
|
|
1SZW
 
 | |
1SU3
 
 | | X-ray structure of human proMMP-1: New insights into collagenase action | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Jozic, D, Bourenkov, G, Lim, N.H, Nagase, H, Bode, W, Maskos, K, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-26 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray structure of human proMMP-1: new insights into procollagenase activation and collagen binding.
J.Biol.Chem., 280, 2005
|
|
1SW8
 
 | | Solution structure of the N-terminal domain of Human N60D calmodulin refined with paramagnetism based strategy | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Bertini, I, Del Bianco, C, Gelis, I, Katsaros, N, Luchinat, C, Parigi, G, Peana, M, Provenzani, A, Zoroddu, M.A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-30 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Experimentally exploring the conformational space sampled by domain reorientation in calmodulin
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TL5
 
 | | Solution structure of apoHAH1 | | Descriptor: | Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1PFJ
 
 | | Solution structure of the N-terminal PH/PTB domain of the TFIIH P62 subunit | | Descriptor: | TFIIH basal transcription factor complex p62 subunit | | Authors: | Gervais, V, Lamour, V, Jawhari, A, Frindel, F, Wasielewski, E, Thierry, J.C, Kieffer, B, Poterszman, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-05-27 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | TFIIH contains a PH domain involved in DNA nucleotide excision repair.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TL4
 
 | | Solution structure of Cu(I) HAH1 | | Descriptor: | COPPER (I) ION, Copper transport protein ATOX1 | | Authors: | Anastassopoulou, I, Banci, L, Bertini, I, Cantini, F, Katsari, E, Rosato, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-09 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Apo and Copper(I)-Loaded Human Metallochaperone HAH1.
Biochemistry, 43, 2004
|
|
1QZZ
 
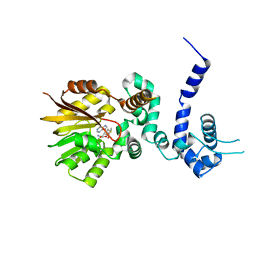 | | Crystal structure of aclacinomycin-10-hydroxylase (RdmB) in complex with S-adenosyl-L-methionine (SAM) | | Descriptor: | ACETATE ION, S-ADENOSYLMETHIONINE, aclacinomycin-10-hydroxylase | | Authors: | Jansson, A, Niemi, J, Lindqvist, Y, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-09-19 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Aclacinomycin-10-Hydroxylase, a S-Adenosyl-L-Methionine-dependent Methyltransferase Homolog Involved in Anthracycline Biosynthesis in Streptomyces purpurascens.
J.Mol.Biol., 334, 2003
|
|
1ZRU
 
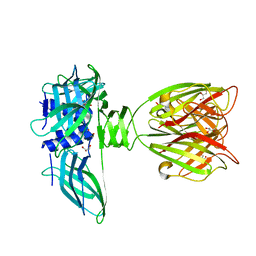 | | structure of the lactophage p2 receptor binding protein in complex with glycerol | | Descriptor: | GLYCEROL, lactophage p2 receptor binding protein | | Authors: | Spinelli, S, Tremblay, D.M, Tegoni, M, Blangy, S, Huyghe, C, Desmyter, A, Labrie, S, de Haard, H, Moineau, S, Cambillau, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-22 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Receptor-binding protein of Lactococcus lactis phages: identification and characterization of the saccharide receptor-binding site.
J.Bacteriol., 188, 2006
|
|
1ZRZ
 
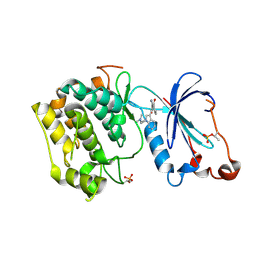 | | Crystal Structure of the Catalytic Domain of Atypical Protein Kinase C-iota | | Descriptor: | 3-{1-[3-(DIMETHYLAMINO)PROPYL]-1H-INDOL-3-YL}-4-(1H-INDOL-3-YL)-1H-PYRROLE-2,5-DIONE, Protein kinase C, iota | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M, Baedeker, M, Benda, C, Jestel, A, Brandstetter, H, Neuefeind, T, Blaesse, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-23 | | Release date: | 2005-09-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Atypical Protein Kinase C-iota Reveals Interaction Mode of Phosphorylation Site in Turn Motif
J.Mol.Biol., 352, 2005
|
|
2OBC
 
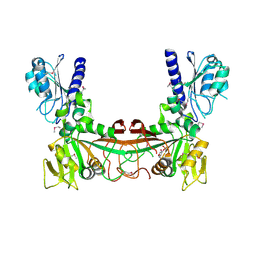 | | The crystal structure of RibD from Escherichia coli in complex with a substrate analogue, ribose 5-phosphate (beta form), bound to the active site of the reductase domain | | Descriptor: | 5-O-phosphono-beta-D-ribofuranose, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-18 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
2O7P
 
 | | The crystal structure of RibD from Escherichia coli in complex with the oxidised NADP+ cofactor in the active site of the reductase domain | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Riboflavin biosynthesis protein ribD | | Authors: | Moche, M, Stenmark, P, Gurmu, D, Nordlund, P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2006-12-11 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of the bifunctional deaminase/reductase RibD of the riboflavin biosynthetic pathway in Escherichia coli: implications for the reductive mechanism.
J.Mol.Biol., 373, 2007
|
|
1SO9
 
 | | Solution Structure of apoCox11, 30 structures | | Descriptor: | Cytochrome C oxidase assembly protein ctaG | | Authors: | Banci, L, Bertini, I, Cantini, F, Ciofi-Baffoni, S, Gonnelli, L, Mangani, S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-13 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cox11, a Novel Type of {beta}-Immunoglobulin-like Fold Involved in CuB Site Formation of Cytochrome c Oxidase.
J.Biol.Chem., 279, 2004
|
|
1SJW
 
 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
1Q0Z
 
 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin A (DcmA) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN A (DCMAA), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-18 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1Q0R
 
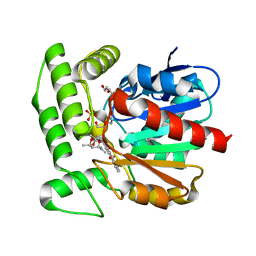 | | Crystal structure of aclacinomycin methylesterase (RdmC) with bound product analogue, 10-decarboxymethylaclacinomycin T (DcmaT) | | Descriptor: | 10-DECARBOXYMETHYLACLACINOMYCIN T (DCMAT), PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Jansson, A, Niemi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-07-17 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of aclacinomycin methylesterase with bound product analogues: implications for anthracycline recognition and mechanism.
J.Biol.Chem., 278, 2003
|
|
1SB6
 
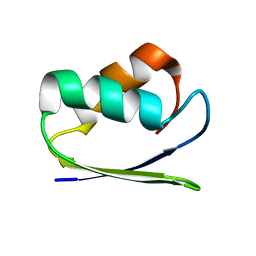 | | Solution structure of a cyanobacterial copper metallochaperone, ScAtx1 | | Descriptor: | copper chaperone ScAtx1 | | Authors: | Banci, L, Bertini, I, Ciofi-Baffoni, S, Su, X.C, Borrelly, G.P, Robinson, N.J, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-02-10 | | Release date: | 2004-04-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of a Cyanobacterial Metallochaperone: INSIGHT INTO AN ATYPICAL COPPER-BINDING MOTIF.
J.Biol.Chem., 279, 2004
|
|
1T5C
 
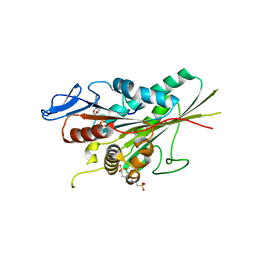 | | Crystal structure of the motor domain of human kinetochore protein CENP-E | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Centromeric protein E, MAGNESIUM ION, ... | | Authors: | Garcia-Saez, I, Yen, T, Wade, R.H, Kozielski, F, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the motor domain of the human kinetochore protein CENP-E.
J.Mol.Biol., 340, 2004
|
|
1TW2
 
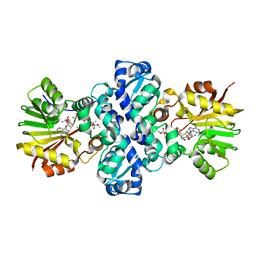 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
2B1U
 
 | | Solution structure of Calmodulin-like Skin Protein C terminal domain | | Descriptor: | Calmodulin-like protein 5 | | Authors: | Babini, E, Bertini, I, Capozzi, F, Chirivino, E, Luchinat, C, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-09-16 | | Release date: | 2006-05-30 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural and Dynamic Characterization of the EF-Hand Protein CLSP.
Structure, 14, 2006
|
|
1YUS
 
 | | Solution structure of apo-S100A13 | | Descriptor: | S100 calcium binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUU
 
 | | Solution structure of Calcium-S100A13 | | Descriptor: | CALCIUM ION, S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
1YUR
 
 | | Solution structure of apo-S100A13 (minimized mean structure) | | Descriptor: | S100 calcium-binding protein A13 | | Authors: | Arnesano, F, Banci, L, Bertini, I, Fantoni, A, Tenori, L, Viezzoli, M.S, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-02-14 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Interplay between Calcium(II) and Copper(II) Binding to S100A13 Protein
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2BZE
 
 | | NMR Structure of human RTF1 PLUS3 domain. | | Descriptor: | KIAA0252 PROTEIN | | Authors: | Truffault, V, Diercks, T, Ab, E, De Jong, R.N, Daniels, M.A, Kaptein, R, Folkers, G.E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-08-16 | | Release date: | 2007-01-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structure and DNA Binding of the Human Rtf1 Plus3 Domain.
Structure, 16, 2008
|
|
1ZT4
 
 | | The crystal structure of human CD1d with and without alpha-Galactosylceramide | | Descriptor: | Beta-2-microglobulin, N-{(1S,2R,3S)-1-[(ALPHA-D-GALACTOPYRANOSYLOXY)METHYL]-2,3-DIHYDROXYHEPTADECYL}HEXACOSANAMIDE, T-cell surface glycoprotein CD1d | | Authors: | Koch, M, Stronge, V.S, Shepherd, D, Gadola, S.D, Mathew, B, Ritter, G, Fersht, A.R, Besra, G.S, Schmidt, R.R, Jones, E.Y, Cerundolo, V, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-26 | | Release date: | 2005-07-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of human CD1d with and without alpha-galactosylceramide
Nat.Immunol., 6, 2005
|
|
