5XQJ
 
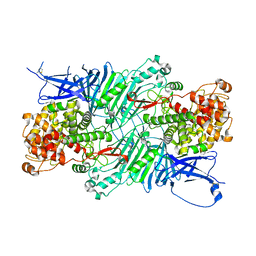 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum complexed with unsaturated galacturonosyl rhamnose substituted with galactose | | Descriptor: | 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-[beta-D-galactopyranose-(1-4)]alpha-L-rhamnopyranose, 2,6-anhydro-3-deoxy-L-threo-hex-2-enonic acid-(1-2)-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-07 | | Release date: | 2018-04-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
5XQ3
 
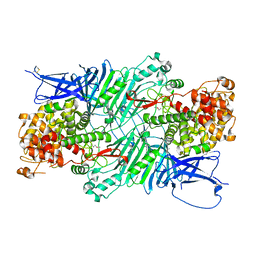 | | Crystal structure of a PL 26 exo-rhamnogalacturonan lyase from Penicillium chrysogenum | | Descriptor: | CALCIUM ION, Pcrglx protein | | Authors: | Kunishige, Y, Iwai, M, Tada, T, Nishimura, S, Sakamoto, T. | | Deposit date: | 2017-06-06 | | Release date: | 2018-03-21 | | Last modified: | 2018-05-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of exo-rhamnogalacturonan lyase from Penicillium chrysogenum as a member of polysaccharide lyase family 26
FEBS Lett., 592, 2018
|
|
8H8N
 
 | | Crystal structure of apo-R52Y/E56Y/R59Y/E63Y-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8L
 
 | | Crystal structure of apo-R52F/E56F/R59F/E63F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8M
 
 | | Crystal structure of apo-E53F/E57F/E60F/E64F-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
8H8O
 
 | | Crystal structure of apo-R52W/E56W/R59W/E63W-rHLFr | | Descriptor: | 1,2-ETHANEDIOL, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Hishikawa, Y, Noya, H, Maity, B, Abe, S, Ueno, T. | | Deposit date: | 2022-10-23 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating Conformational Dynamics and Thermostability of Designed Aromatic Clusters by Using Protein Cages.
Chemistry, 29, 2023
|
|
6IOS
 
 | | The ligand binding domain of Mlp24 with proline | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Calcium Ions Modulate Amino Acid Sensing of the Chemoreceptor Mlp24 ofVibrio cholerae.
J. Bacteriol., 201, 2019
|
|
6IOV
 
 | | The ligand binding domain of Mlp37 with arginine | | Descriptor: | ARGININE, Methyl-accepting chemotaxis (MCP) signaling domain protein | | Authors: | Takahashi, Y, Sumita, K, Nishiyama, S, Kawagishi, I, Imada, K. | | Deposit date: | 2018-10-31 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Structural basis of the binding affinity of chemoreceptors Mlp24p and Mlp37p for various amino acids.
Biochem.Biophys.Res.Commun., 523, 2020
|
|
5XSV
 
 | |
5XSX
 
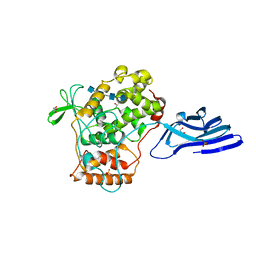 | | Crystal structure of an archaeal chitinase in the substrate-complex form (P212121) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase, GLYCEROL, ... | | Authors: | Nishitani, Y, Miki, K. | | Deposit date: | 2017-06-15 | | Release date: | 2018-05-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Crystal structures of an archaeal chitinase ChiD and its ligand complexes.
Glycobiology, 28, 2018
|
|
4YN5
 
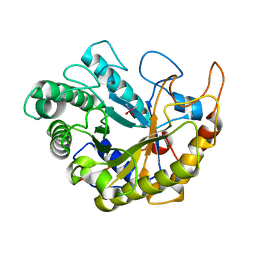 | | Catalytic domain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase | | Descriptor: | CACODYLATE ION, Mannan endo-1,4-beta-mannosidase | | Authors: | Shimane, Y, Ohta, Y, Usami, R, Hatada, Y. | | Deposit date: | 2015-03-09 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase
To Be Published
|
|
8GQ9
 
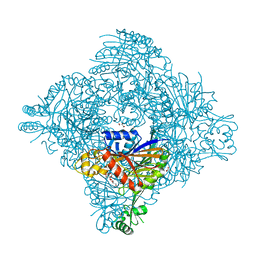 | | Crystal structure of lasso peptide epimerase MslH | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nakashima, Y, Morita, H. | | Deposit date: | 2022-08-29 | | Release date: | 2023-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism.
Nat Commun, 14, 2023
|
|
8GQB
 
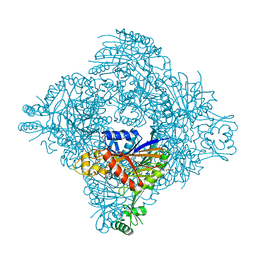 | |
8GQA
 
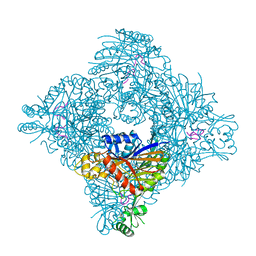 | |
7A1O
 
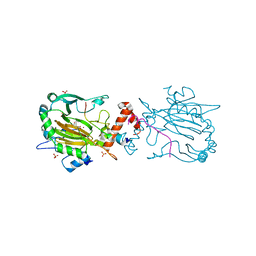 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 4-ethyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 4-ethyl-2-oxoglutarate, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1Q
 
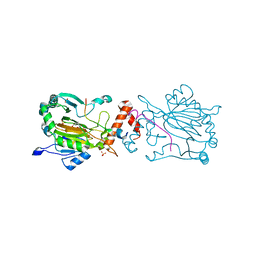 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | 3-(carboxycarbonyl)cyclopentane-1-carboxylic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1P
 
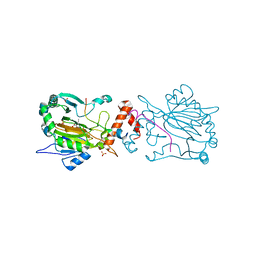 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 4-propyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | (4~{S})-2-oxidanylidene-4-propyl-pentanedioic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1M
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-propyl-2-oxoglutarate | | Descriptor: | 3-propyl-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1N
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-methyl-2-oxoglutarate, AND CONSENSUS ANKYRIN REPEAT DOMAIN (20-MER) | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, CONSENSUS ANKYRIN REPEAT DOMAIN, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1L
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-methyl-2-oxoglutarate | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1J
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-(3-phenylpropyl)-2-oxoglutarate | | Descriptor: | 3-(3-phenylpropyl)-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1S
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II), 3-methyl-2-oxoglutarate, AND TANKYRASE-2 (TNKS2) FRAGMENT PEPTIDE (21-MER) | | Descriptor: | (3~{S})-3-methyl-2-oxidanylidene-pentanedioic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
7A1K
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH ZN(II) AND 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate | | Descriptor: | 3-((9,9-dimethyl-9H-fluoren-2-yl)methyl)-2-oxoglutarate, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Brewitz, L, Schofield, C.J. | | Deposit date: | 2020-08-13 | | Release date: | 2021-08-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | 2-Oxoglutarate derivatives can selectively enhance or inhibit the activity of human oxygenases.
Nat Commun, 12, 2021
|
|
5KC1
 
 | | Structure of the C-terminal dimerization domain of Atg38 | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Autophagy-related protein 38, ... | | Authors: | Ohashi, Y, Soler, N, Garcia-Ortegon, M, Zhang, L, Perisic, O, Masson, G.R, Johnson, C.M, Williams, R.J. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
7R8I
 
 | |
