5NUS
 
 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | Descriptor: | ZINC ION, p34, p44 | | Authors: | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | Deposit date: | 2017-05-02 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU3
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU2
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU5
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
1Q51
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB in Complex with Acetoacetyl-Coenzyme A, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | ACETOACETYL-COENZYME A, menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1Q52
 
 | | Crystal Structure of Mycobacterium tuberculosis MenB, a Key Enzyme in Vitamin K2 Biosynthesis | | Descriptor: | menB | | Authors: | Truglio, J.J, Theis, K, Feng, Y, Gajda, R, Machutta, C, Tonge, P.J, Kisker, C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-08-05 | | Release date: | 2004-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
J.Biol.Chem., 278, 2003
|
|
1QRE
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRL
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | BICARBONATE ION, CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRF
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION, SULFATE ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRM
 
 | | A CLOSER LOOK AT THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTAL STRUCTURES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, SULFATE ION, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QQ0
 
 | | COBALT SUBSTITUTED CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, COBALT (II) ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-10 | | Release date: | 1999-06-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
1QRG
 
 | | A CLOSER LOOK AND THE ACTIVE SITE OF GAMMA-CARBONIC ANHYDRASES: HIGH RESOLUTION CRYSTALLOGRAPHIC STUDIES OF THE CARBONIC ANHYDRASE FROM METHANOSARCINA THERMOPHILA | | Descriptor: | CARBONIC ANHYDRASE, ZINC ION | | Authors: | Iverson, T.M, Alber, B.E, Kisker, C, Ferry, J.G, Rees, D.C. | | Deposit date: | 1999-06-13 | | Release date: | 1999-06-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A closer look at the active site of gamma-class carbonic anhydrases: high-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila.
Biochemistry, 39, 2000
|
|
5ONC
 
 | | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis is Controlled by a Redox-Sensitive Thiol Switch | | Descriptor: | CHLORIDE ION, Steroid 3-ketoacyl-CoA thiolase | | Authors: | Schaefer, C, Kuper, J, Sampson, N.S, Kisker, C. | | Deposit date: | 2017-08-03 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Catabolism of the Cholesterol Side Chain in Mycobacterium tuberculosis Is Controlled by a Redox-Sensitive Thiol Switch.
ACS Infect Dis, 3, 2017
|
|
6EX8
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-~{N}-[1-(aminomethyl)cyclopropyl]-19-chloranyl-5-oxidanylidene-9-(trifluoromethyl)-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6EXQ
 
 | | Crystal Structure of Rhodesain in complex with a Macrolactam Inhibitor | | Descriptor: | (3~{S})-19-chloranyl-~{N}-(1-cyanocyclopropyl)-8-methoxy-5-oxidanylidene-12,17-dioxa-4-azatricyclo[16.2.2.0^{6,11}]docosa-1(20),6(11),7,9,18,21-hexaene-3-carboxamide, 1,2-ETHANEDIOL, Cysteine protease | | Authors: | Dietzel, U, Kisker, C. | | Deposit date: | 2017-11-08 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Repurposing a Library of Human Cathepsin L Ligands: Identification of Macrocyclic Lactams as Potent Rhodesain and Trypanosoma brucei Inhibitors.
J. Med. Chem., 61, 2018
|
|
6H4J
 
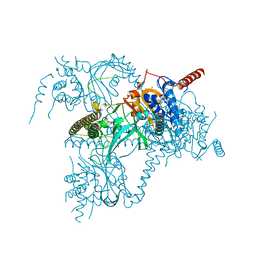 | | Usp25 catalytic domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4I
 
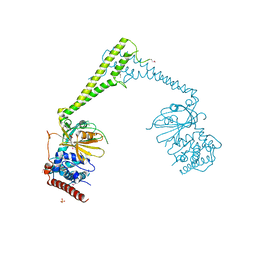 | | Usp28 catalytic domain apo | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4H
 
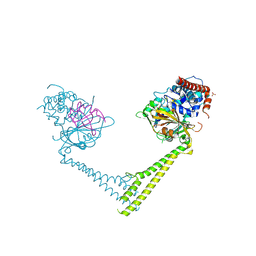 | | Usp28 catalytic domain variant E593D in complex with UbPA | | Descriptor: | Polyubiquitin-B, SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 28, ... | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
6H4K
 
 | | Structure of the Usp25 C-terminal domain | | Descriptor: | CHLORIDE ION, Ubiquitin carboxyl-terminal hydrolase 25 | | Authors: | Klemm, T.A, Sauer, F, Kisker, C. | | Deposit date: | 2018-07-21 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Differential Oligomerization of the Deubiquitinases USP25 and USP28 Regulates Their Activities.
Mol.Cell, 74, 2019
|
|
2W3R
 
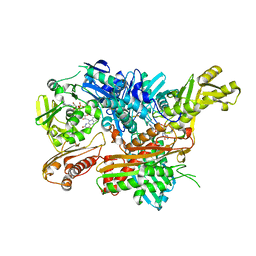 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2A99
 
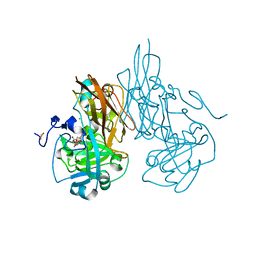 | | Crystal structure of recombinant chicken sulfite oxidase at resting state | | Descriptor: | CHLORIDE ION, GLYCEROL, MOLYBDENUM ATOM, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2A9A
 
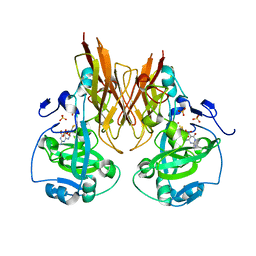 | | Crystal structure of recombinant chicken sulfite oxidase with the bound product, sulfate, at the active site | | Descriptor: | MOLYBDENUM ATOM, PHOSPHONIC ACIDMONO-(2-AMINO-5,6-DIMERCAPTO-4-OXO-3,7,8A,9,10,10A-HEXAHYDRO-4H-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-7-YLMETHYL)ESTER, SULFATE ION, ... | | Authors: | Karakas, E, Wilson, H.L, Graf, T.N, Xiang, S, Jaramillo-Busquets, S, Rajagopalan, K.V, Kisker, C. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structural insights into sulfite oxidase deficiency
J.Biol.Chem., 280, 2005
|
|
2W54
 
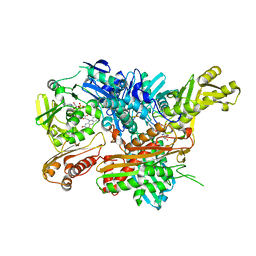 | | Crystal Structure of Xanthine Dehydrogenase from Rhodobacter capsulatus in Complex with Bound Inhibitor Pterin-6-aldehyde | | Descriptor: | 6-HYDROXYMETHYLPTERIN, BARIUM ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Doebbler, J.A, Truglio, J.J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-12-04 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
2WGG
 
 | |
