1NZR
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
7XHA
 
 | |
1OGS
 
 | | human acid-beta-glucosidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Dvir, H, Harel, M, McCarthy, A.A, Toker, L, Silman, I, Futerman, A.H, Sussman, J.L. | | Deposit date: | 2003-05-13 | | Release date: | 2003-07-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structure of Human Acid-Beta-Glucosidase, the Defective Enzyme in Gaucher Disease
Embo Rep., 4, 2003
|
|
7XHB
 
 | |
1OKV
 
 | | Cyclin A binding groove inhibitor H-Arg-Arg-Leu-Ile-Phe-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, H-ARG-ARG-LEU-ILE-PHE-NH2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-07-30 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1LBK
 
 | | Crystal structure of a recombinant glutathione transferase, created by replacing the last seven residues of each subunit of the human class pi isoenzyme with the additional C-terminal helix of human class alpha isoenzyme | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLUTATHIONE, Glutathione S-transferase class pi chimaera (CODA), ... | | Authors: | Kong, G.K.W, Micaloni, C, Mazzetti, A.P, Nuccetelli, M, Antonini, G, Stella, L, McKinstry, W.J, Polekhina, G, Rossjohn, J, Federici, G, Ricci, G, Parker, M.W, Lo Bello, M. | | Deposit date: | 2002-04-04 | | Release date: | 2002-04-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Engineering a new C-terminal tail in the H-site of human glutathione transferase P1-1: structural and functional consequences.
J.Mol.Biol., 325, 2003
|
|
1LN0
 
 | | Structure of the Catalytic Domain of Homing Endonuclease I-TevI | | Descriptor: | SULFATE ION, intron-associated endonuclease 1 | | Authors: | Van Roey, P, Meehan, L, Kowalski, J.C, Belfort, M, Derbyshire, V. | | Deposit date: | 2002-05-02 | | Release date: | 2002-10-30 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI.
Nat.Struct.Biol., 9, 2002
|
|
1OGQ
 
 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OIV
 
 | | X-ray structure of the small G protein Rab11a in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5'-DIPHOSPHATE, RAS-RELATED PROTEIN RAB-11A, ... | | Authors: | Pasqualato, S, Senic-Matuglia, F, Renault, L, Goud, B, Salamero, J, Cherfils, J. | | Deposit date: | 2003-06-26 | | Release date: | 2004-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Structural Gdp/GTP Cycle of Rab11 Reveals a Novel Interface Involved in the Dynamics of Recycling Endosomes
J.Biol.Chem., 279, 2004
|
|
1OJT
 
 | |
5VII
 
 | | Crystal structure of GluN1/GluN2A NMDA receptor agonist binding domains with glycine and antagonist, 4-(3-fluoropropyl)phenyl-ACEPC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(2R)-2-amino-2-carboxyethyl]-1-[4-(3-fluoropropyl)phenyl]-1H-pyrazole-3-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Mou, T.-C, Conti, P, Pinto, A, Tamborini, L, Sprang, S.R, Hansen, K.B. | | Deposit date: | 2017-04-16 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structural basis of subunit selectivity for competitive NMDA receptor antagonists with preference for GluN2A over GluN2B subunits.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VG9
 
 | | Structure of the eukaryotic intramembrane Ras methyltransferase ICMT (isoprenylcysteine carboxyl methyltransferase) without a monobody | | Descriptor: | Protein-S-isoprenylcysteine O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Long, S.B, Diver, M.M, Pedi, L, Koide, A, Koide, S. | | Deposit date: | 2017-04-10 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Atomic structure of the eukaryotic intramembrane RAS methyltransferase ICMT.
Nature, 553, 2018
|
|
1JNL
 
 | | Crystal Structure of Fab-Estradiol Complexes | | Descriptor: | monoclonal anti-estradiol 17E12E5 immunoglobulin gamma-1 chain, monoclonal anti-estradiol 17E12E5 immunoglobulin kappa chain | | Authors: | Monnet, C, Bettsworth, F, Stura, E.A, Le Du, M.-H, Menez, R, Derrien, L, Zinn-Justin, S, Gilquin, B, Sibai, G, Battail-Poirot, N, Jolivet, M, Menez, A, Arnaud, M, Ducancel, F, Charbonnier, J.B. | | Deposit date: | 2001-07-24 | | Release date: | 2002-02-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Highly specific anti-estradiol antibodies: structural characterisation and binding diversity.
J.Mol.Biol., 315, 2002
|
|
1JIH
 
 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
1JLF
 
 | | CRYSTAL STRUCTURE OF Y188C MUTANT HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 RT A-chain, HIV-1 RT B-chain | | Authors: | Ren, J, Nichols, C, Bird, L, Chamberlain, P, Weaver, K, Short, S, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors.
J.Mol.Biol., 312, 2001
|
|
1Q8Q
 
 | | Pterocarpus angolensis lectin (PAL) in complex with the dimannoside Man(alpha1-4)Man | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, alpha-D-mannopyranose-(1-4)-methyl alpha-D-mannopyranoside, ... | | Authors: | Loris, R, Van Walle, I, De Greve, H, Beeckmans, S, Deboeck, F, Wyns, L, Bouckaert, J. | | Deposit date: | 2003-08-22 | | Release date: | 2004-02-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Basis of Oligomannose Recognition by the Pterocarpus angolensis Seed Lectin
J.Mol.Biol., 335, 2004
|
|
5V5X
 
 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1Q94
 
 | | Structures of HLA-A*1101 in complex with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle anchor residue | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-22 | | Release date: | 2004-06-01 | | Last modified: | 2020-02-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
1QKW
 
 | | Alpha-spectrin Src Homology 3 domain, N47G mutant in the distal loop. | | Descriptor: | ALPHA II SPECTRIN, GLYCEROL, SULFATE ION | | Authors: | Vega, M.C, Martinez, J, Serrano, L. | | Deposit date: | 1999-08-16 | | Release date: | 2000-08-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Thermodynamic and structural characterization of Asn and Ala residues in the disallowed II' region of the Ramachandran plot.
Protein Sci., 9, 2000
|
|
1JUB
 
 | | The K136E mutant of lactococcus lactis dihydroorotate dehydrogenase A | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Norager, S, Arent, S, Bjornberg, O, Ottosen, M, Lo Leggio, L, Jensen, K.F, Larsen, S. | | Deposit date: | 2001-08-24 | | Release date: | 2003-09-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Lactococcus lactis dihydroorotate dehydrogenase A mutants reveal important facets of the enzymatic function
J.Biol.Chem., 278, 2003
|
|
5UP1
 
 | | Solution structure of the de novo mini protein EEHEE_rd3_1049 | | Descriptor: | EEHEE_rd3_1049 | | Authors: | Houliston, S, Rocklin, G.J, Lemak, A, Carter, L, Chidyausiku, T.M, Baker, D, Arrowsmith, C.H. | | Deposit date: | 2017-02-01 | | Release date: | 2017-07-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Global analysis of protein folding using massively parallel design, synthesis, and testing.
Science, 357, 2017
|
|
1JVK
 
 | | THREE-DIMENSIONAL STRUCTURE OF AN IMMUNOGLOBULIN LIGHT CHAIN DIMER ACTING AS A LETHAL AMYLOID PRECURSOR | | Descriptor: | IMMUNOGLOBULIN LAMBDA LIGHT CHAIN | | Authors: | Bourne, P.C, Ramsland, P.A, Shan, L, Fan, Z.-C, DeWitt, C.R, Shultz, B.B, Terzyan, S.S, Edmundson, A.B. | | Deposit date: | 2001-08-30 | | Release date: | 2002-05-03 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1QI7
 
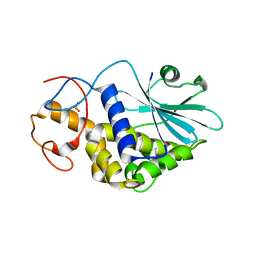 | | THE CRYSTAL STRUCTURE AT 2.0 A OF SAPORIN SO6, A RIBOSOME INACTIVATING PROTEIN FROM SAPONARIA OFFICINALIS | | Descriptor: | PROTEIN (N-GLYCOSIDASE), SULFATE ION | | Authors: | Savino, C, Federici, L, Ippoliti, R, Lendaro, E, Tsernoglou, D. | | Deposit date: | 1999-06-08 | | Release date: | 2000-06-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of saporin SO6 from Saponaria officinalis and its interaction with the ribosome.
FEBS Lett., 470, 2000
|
|
5VFB
 
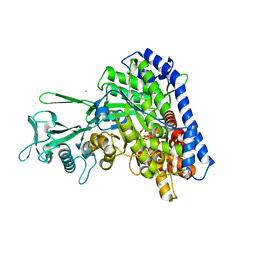 | | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid. | | Descriptor: | CHLORIDE ION, GLYCOLIC ACID, Malate synthase G, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-07 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | 1.36 Angstrom Resolution Crystal Structure of Malate Synthase G from Pseudomonas aeruginosa in Complex with Glycolic Acid.
To Be Published
|
|
1K0V
 
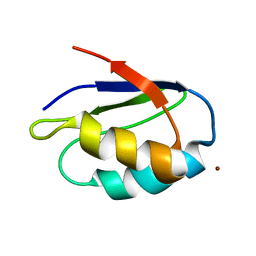 | | Copper trafficking: the solution structure of Bacillus subtilis CopZ | | Descriptor: | COPPER (I) ION, CopZ | | Authors: | Banci, L, Bertini, I, Del Conte, R, Markey, J, Ruiz-Duenas, F.J. | | Deposit date: | 2001-09-21 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Copper trafficking: the solution structure of Bacillus subtilis CopZ.
Biochemistry, 40, 2001
|
|
