4RD7
 
 | | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205 | | Descriptor: | Cupin 2 conserved barrel domain protein, GLYCEROL, SULFATE ION | | Authors: | Tan, K, Gu, M, Clancy, S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | The crystal structure of a Cupin 2 conserved barrel domain protein from Salinispora arenicola CNS-205
To be Published
|
|
3NYM
 
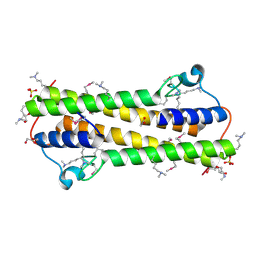 | | The crystal structure of functionally unknown protein from Neisseria meningitidis MC58 | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Zhang, R, Tan, K, Volkart, L, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-15 | | Release date: | 2010-09-22 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of functionally unknown protein from Neisseria meningitidis MC58
To be Published
|
|
3D3Y
 
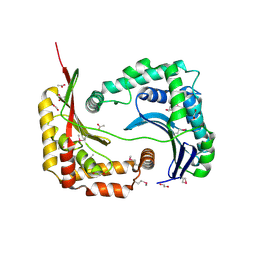 | | Crystal structure of a conserved protein from Enterococcus faecalis V583 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Uncharacterized protein | | Authors: | Tan, K, Li, H, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-13 | | Release date: | 2008-07-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of a conserved protein from Enterococcus faecalis V583.
To be Published
|
|
4U12
 
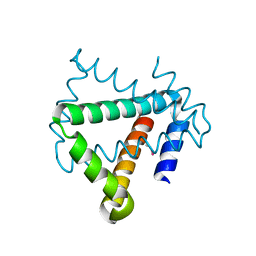 | | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer | | Descriptor: | Uncharacterized protein HP0242 | | Authors: | Grabowski, M, Shabalin, I.G, Chruszcz, M, Skarina, T, Onopriyenko, O, Guthrie, J, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-14 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of protein HP0242 from Helicobacter pylori at 1.94 A resolution: a knotted homodimer
to be published
|
|
4U28
 
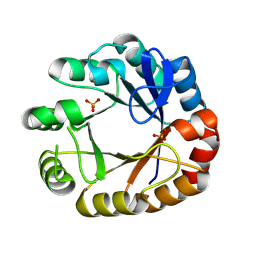 | | Crystal structure of apo Phosphoribosyl isomerase A from Streptomyces sviceus ATCC 29083 | | Descriptor: | PHOSPHATE ION, Phosphoribosyl isomerase A | | Authors: | Chang, C, Verduzco-Castro, E.A, Endres, M, Barona-Gomez, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Co-occurrence of analogous enzymes determines evolution of a novel ( beta alpha )8-isomerase sub-family after non-conserved mutations in flexible loop.
Biochem. J., 473, 2016
|
|
3C5P
 
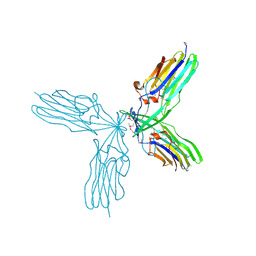 | | Crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, Protein BAS0735 of unknown function | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne.
To be Published
|
|
4EYG
 
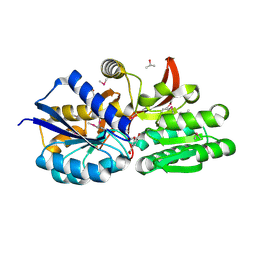 | | Crystal structure of solute binding protein of ABC transporter from Rhodopseudomonas palustris BisB5 in complex with vanillic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXY-3-METHOXYBENZOATE, ISOPROPYL ALCOHOL, ... | | Authors: | Chang, C, Mack, J, Zerbs, S, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-05-01 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Characterization of transport proteins for aromatic compounds derived from lignin: benzoate derivative binding proteins.
J.Mol.Biol., 423, 2012
|
|
4HD1
 
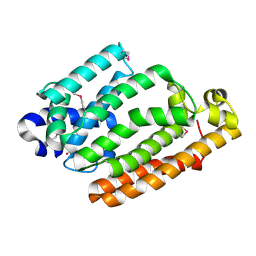 | |
1Y88
 
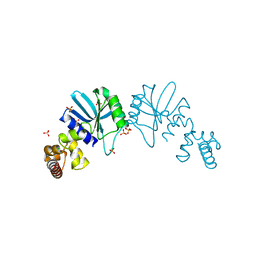 | | Crystal Structure of Protein of Unknown Function AF1548 | | Descriptor: | CHLORIDE ION, Hypothetical protein AF1548, SULFATE ION | | Authors: | Lunin, V.V, Evdokimova, E, Kudritskaya, M, Cuff, M.E, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-12-10 | | Release date: | 2004-12-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of hypothetical protein AF1548 from Archaeoglobus fulgidus
To be Published
|
|
4HN9
 
 | |
3CNI
 
 | | Crystal structure of a domain of a putative ABC type-2 transporter from Thermotoga maritima MSB8 | | Descriptor: | CALCIUM ION, Putative ABC type-2 transporter | | Authors: | Filippova, E.V, Shumilin, I, Tkaczuk, K.L, Cymborowski, M, Chruszcz, M, Xu, X, Que, Q, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-25 | | Release date: | 2008-04-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the putative ABC-type 2 transporter from Thermotoga maritima MSB8.
J.Struct.Funct.Genom., 15, 2014
|
|
4KV7
 
 | |
1G60
 
 | | Crystal Structure of Methyltransferase MboIIa (Moraxella bovis) | | Descriptor: | Adenine-specific Methyltransferase MboIIA, S-ADENOSYLMETHIONINE, SODIUM ION | | Authors: | Osipiuk, J, Walsh, M.A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-11-02 | | Release date: | 2002-05-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of MboIIA methyltransferase.
Nucleic Acids Res., 31, 2003
|
|
3H0K
 
 | |
3KBQ
 
 | |
4PW0
 
 | |
3HCZ
 
 | | The crystal structure of a domain of possible thiol-disulfide isomerase from Cytophaga hutchinsonii ATCC 33406. | | Descriptor: | FORMIC ACID, Possible thiol-disulfide isomerase, SULFATE ION | | Authors: | Tan, K, Marshall, N, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-06 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | The crystal structure of a domain of possible thiol-disulfide isomerase from Cytophaga hutchinsonii ATCC 33406.
To be Published
|
|
4EMY
 
 | |
3KBR
 
 | | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Cyclohexadienyl dehydratase, ... | | Authors: | Tan, K, Marshall, N, Buck, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | The crystal structure of cyclohexadienyl dehydratase precursor from Pseudomonas aeruginosa PA01
To be Published
|
|
3KK4
 
 | | uncharacterized protein BP1543 from Bordetella pertussis Tohama I | | Descriptor: | ACETATE ION, GLYCEROL, uncharacterized protein BP1543 | | Authors: | Chang, C, Chhor, G, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-04 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of uncharacterized protein BP1543 from Bordetella pertussis Tohama I
To be Published
|
|
3KKC
 
 | | The crystal structure OF TetR transcriptional regulator from Streptococcus agalactiae 2603V | | Descriptor: | IMIDAZOLE, NICKEL (II) ION, TetR family Transcriptional regulator | | Authors: | Tan, K, Hatzos, C, Morgan, T, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-05 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure OF TetR transcriptional regulator from Streptococcus agalactiae 2603V
To be Published
|
|
3HDT
 
 | | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940 | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, putative kinase | | Authors: | Chang, C, Freeman, L, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-07 | | Release date: | 2009-05-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Crystal structure of putative kinase from Clostridium symbiosum ATCC 14940
To be Published
|
|
4HVM
 
 | | Crystal structure of tallysomycin biosynthesis protein TlmII | | Descriptor: | SULFATE ION, TlmII | | Authors: | Chang, C, Bigelow, L, Bearden, J, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-06 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Crystal structure of tallysomycin biosynthesis protein TlmII
To be Published
|
|
3HJZ
 
 | | The structure of an aldolase from Prochlorococcus marinus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Xu, X, Cui, H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-05-22 | | Release date: | 2009-06-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3KN3
 
 | | Crystal Structure of LysR Substrate Binding Domain (25-263) of Putative Periplasmic Protein from Wolinella succinogenes | | Descriptor: | ACETIC ACID, CITRIC ACID, GLUTATHIONE, ... | | Authors: | Kim, Y, Volkart, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-11 | | Release date: | 2009-12-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.412 Å) | | Cite: | Crystal Structure of LysR Substrate Binding Domain from Wolinella succinogenes
To be Published
|
|
