3C8H
 
 | | Crystal structure of the enterobactin esterase FES from Shigella flexneri in the presence of 2,3-Di-hydroxy-N-benzoyl-serine | | Descriptor: | Enterochelin esterase | | Authors: | Kim, Y, Maltseva, N, Abergel, R, Holzle, D, Raymond, K, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-12 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Siderophore Mediated Iron Acquisition: Structure and Specificity of Enterobactin Esterase from Shigella flexneri.
To be Published
|
|
1H55
 
 | | STRUCTURE OF HORSERADISH PEROXIDASE C1A COMPOUND II | | Descriptor: | ACETATE ION, CALCIUM ION, OXYGEN ATOM, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-18 | | Release date: | 2002-06-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1H5K
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (78-89% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1QX9
 
 | |
1QL4
 
 | | Structure of the soluble domain of cytochrome c552 from Paracoccus denitrificans in the oxidised state | | Descriptor: | CYTOCHROME C552, HEME C | | Authors: | Harrenga, A, Reincke, B, Rueterjans, H, Ludwig, B, Michel, H. | | Deposit date: | 1999-08-20 | | Release date: | 2000-02-03 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Soluble Domain of Cytochrome C552 from Paracoccus Denitrificans in the Oxidized and Reduced States
J.Mol.Biol., 295, 2000
|
|
3BUU
 
 | |
1H5E
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (11-22% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
7ASK
 
 | | 43S preinitiation complex from Trypanosoma cruzi with the kDDX60 helicase bound with ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, kDDX60 | | Authors: | Bochler, A, Brito Querido, J, Prilepskaja, T, Soufari, H, Del Cistia, M.L, Kuhn, L, Rimoldi Ribeiro, A, Valasek, L.S, Hashem, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-14 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Cell Rep, 33, 2020
|
|
3C1G
 
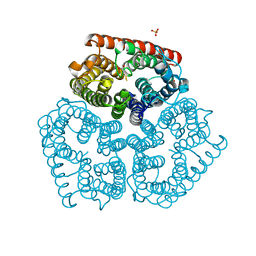 | | Substrate binding, deprotonation and selectivity at the periplasmic entrance of the E. coli ammonia channel AmtB | | Descriptor: | ACETATE ION, Ammonia channel, SULFATE ION | | Authors: | Javelle, A, Kostrewa, D, Lupo, D, Winkler, F.K. | | Deposit date: | 2008-01-23 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1QTT
 
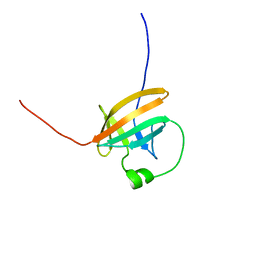 | | SOLUTION STRUCTURE OF THE ONCOPROTEIN P13MTCP1 | | Descriptor: | PRODUCT OF THE MTCP1 ONCOGENE | | Authors: | Guignard, L, Padilla, A, Mispelter, J, Yang, Y.-S, Stern, M.-H, Lhoste, J.-M, Roumestand, C. | | Deposit date: | 1999-06-29 | | Release date: | 2001-01-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and solution structure refinement of the 15N-labeled human oncogenic protein p13MTCP1: comparison with X-ray data.
J.Biomol.NMR, 17, 2000
|
|
7ASE
 
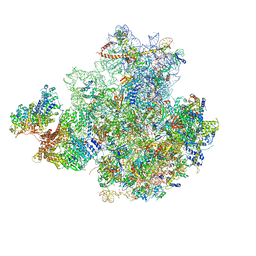 | | 43S preinitiation complex from Trypanosoma cruzi with the kDDX60 helicase | | Descriptor: | 18S, 40S ribosomal protein S10, putative, ... | | Authors: | Bochler, A, Brito Querido, J, Prilepskaja, T, Soufari, H, Del Cistia, M.L, Kuhn, L, Rimoldi Ribeiro, A, Valasek, L.S, Hashem, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural Differences in Translation Initiation between Pathogenic Trypanosomatids and Their Mammalian Hosts.
Cell Rep, 33, 2020
|
|
3BL3
 
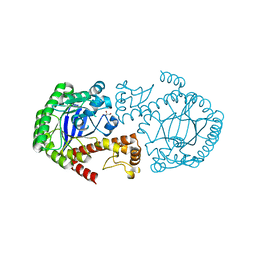 | | tRNA guanine transglycosylase V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-10 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
1H5G
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (33-44% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
3BQW
 
 | |
1QFR
 
 | | NMR SOLUTION STRUCTURE OF PHOSPHOCARRIER PROTEIN HPR FROM ENTEROCOCCUS FAECALIS | | Descriptor: | PHOSPHOCARRIER PROTEIN HPR | | Authors: | Maurer, T, Doeker, R, Goerler, A, Hengstenberg, W, Kalbitzer, H.R. | | Deposit date: | 1999-04-13 | | Release date: | 2001-02-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of the histidine-containing phosphocarrier protein (HPr) from Enterococcus faecalis in solution.
Eur.J.Biochem., 268, 2001
|
|
3C5P
 
 | | Crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne | | Descriptor: | MAGNESIUM ION, Protein BAS0735 of unknown function | | Authors: | Kim, Y, Joachimiak, G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-01 | | Release date: | 2008-02-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of BAS0735, a protein of unknown function from Bacillus anthracis str. Sterne.
To be Published
|
|
1QA1
 
 | | TAILSPIKE PROTEIN, MUTANT V331G | | Descriptor: | TAILSPIKE PROTEIN | | Authors: | Baxa, U, Steinbacher, S, Weintraub, A, Huber, R, Seckler, R. | | Deposit date: | 1999-04-10 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mutations improving the folding of phage P22 tailspike protein affect its receptor binding activity.
J.Mol.Biol., 293, 1999
|
|
1HF6
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE | | Descriptor: | ACETIC ACID, ENDOGLUCANASE B, GLYCEROL, ... | | Authors: | Varrot, A, Withers, S, Vasella, A, Schulein, M, Davies, G.J. | | Deposit date: | 2000-11-29 | | Release date: | 2001-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Direct Experimental Observation of the Hydrogen-Bonding Network of a Glycosidase Along its Reaction Coordinate Revealed by Atomic Resolution Analyses of Endoglucanase Cel5A
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
1H36
 
 | | Structures of Human Oxidosqualene Cyclase Inhibitors Bound to an Homologous Enzyme | | Descriptor: | (4-BROMOPHENYL)[4-({(2E)-4-[CYCLOPROPYL(METHYL)AMINO]BUT-2-ENYL}OXY)PHENYL]METHANONE, (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, SQUALENE--HOPENE CYCLASE | | Authors: | Lenhart, A, Reinert, D.J, Weihofen, W.A, Aebi, J.D, Dehmlow, H, Morand, O.H, Schulz, G.E. | | Deposit date: | 2002-08-24 | | Release date: | 2003-08-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structures and Potencies of Oxidosqualene Cyclase Inhibitors with the Homologous Squalene-Hopene Cyclase
J.Med.Chem., 46, 2003
|
|
3C7C
 
 | | A structural basis for substrate and stereo selectivity in octopine dehydrogenase (ODH-NADH-L-Arginine) | | Descriptor: | ARGININE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Octopine dehydrogenase | | Authors: | Smits, S.H.J, Mueller, A, Schmitt, L, Grieshaber, M.K. | | Deposit date: | 2008-02-07 | | Release date: | 2008-07-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A structural basis for substrate selectivity and stereoselectivity in octopine dehydrogenase from Pecten maximus.
J.Mol.Biol., 381, 2008
|
|
1H5H
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (44-56% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, HYDROGEN PEROXIDE, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-22 | | Release date: | 2002-06-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1QSH
 
 | | MAGNESIUM(II)-AND ZINC(II)-PROTOPORPHYRIN IX'S STABILIZE THE LOWEST OXYGEN AFFINITY STATE OF HUMAN HEMOGLOBIN EVEN MORE STRONGLY THAN DEOXYHEME | | Descriptor: | PROTEIN (HEMOGLOBIN ALPHA CHAIN), PROTEIN (HEMOGLOBIN BETA CHAIN), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Miyazaki, G, Morimoto, H, Yun, K.-M, Park, S.-Y, Nakagawa, A, Minagawa, H, Shibayama, N. | | Deposit date: | 1999-06-22 | | Release date: | 1999-07-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Magnesium(II) and zinc(II)-protoporphyrin IX's stabilize the lowest oxygen affinity state of human hemoglobin even more strongly than deoxyheme.
J.Mol.Biol., 292, 1999
|
|
1H5D
 
 | | X-ray induced reduction of horseradish peroxidase C1A Compound III (0-11% dose) | | Descriptor: | ACETATE ION, CALCIUM ION, PEROXIDASE C1A, ... | | Authors: | Berglund, G.I, Carlsson, G.H, Hajdu, J, Smith, A.T, Szoke, H, Henriksen, A. | | Deposit date: | 2001-05-21 | | Release date: | 2002-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Catalytic Pathway of Horseradish Peroxidase at High Resolution
Nature, 417, 2002
|
|
1QQU
 
 | |
