4Q29
 
 | | Ensemble Refinement of plu4264 protein from Photorhabdus luminescens | | Descriptor: | NICKEL (II) ION, SODIUM ION, plu4264 protein | | Authors: | Wang, F, Michalska, K, Li, H, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R, Weerth, S, Miller, M.D, Thomas, M.G, Joachimiak, A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-04-07 | | Release date: | 2014-05-07 | | Last modified: | 2015-02-11 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Structure of a cupin protein Plu4264 from Photorhabdus luminescens subsp. laumondii TTO1 at 1.35 angstrom resolution.
Proteins, 83, 2015
|
|
4Q31
 
 | | The crystal structure of cystathione gamma lyase (CalE6) from Micromonospora echinospora | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Tan, K, Bigelow, L, Jedrzejczak, R, Babnigg, G, Bingman, C.A, Yennamalli, R.M, Singh, S, Kharel, M.K, Thorson, J.S, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-04-10 | | Release date: | 2014-05-07 | | Last modified: | 2017-03-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural dynamics of a methionine gamma-lyase for calicheamicin biosynthesis: Rotation of the conserved tyrosine stacking with pyridoxal phosphate.
Struct Dyn, 3, 2016
|
|
4QA9
 
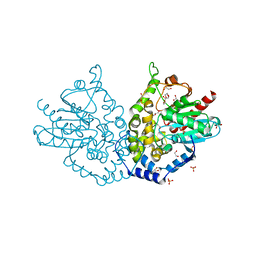 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4F4I
 
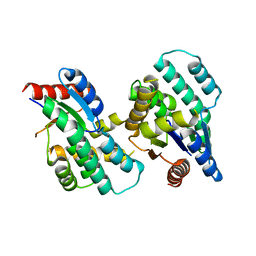 | | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form | | Descriptor: | Thymidylate kinase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Jedrzejczak, R, Babnigg, G, Rubin, E, Sacchettini, J, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-05-10 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of Thymidylate Kinase from Staphylococcus aureus in apo-form
To be Published
|
|
4IAG
 
 | | Crystal structure of ZbmA, the zorbamycin binding protein from Streptomyces flavoviridis | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Zbm binding protein | | Authors: | Cuff, M.E, Bigelow, L, Bruno, C.J.P, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-12-06 | | Release date: | 2013-02-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
1XBW
 
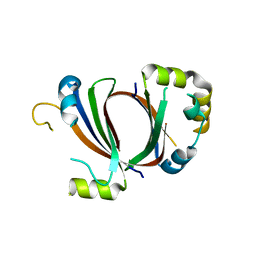 | | 1.9A Crystal Structure of the protein isdG from Staphylococcus aureus aureus, Structural genomics, MCSG | | Descriptor: | hypothetical protein isdG | | Authors: | Zhang, R, Wu, R, Joachimiak, G, Schneewind, O, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-31 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Staphylococcus aureus IsdG and IsdI, heme-degrading enzymes with structural similarity to monooxygenases.
J.Biol.Chem., 280, 2005
|
|
1XDZ
 
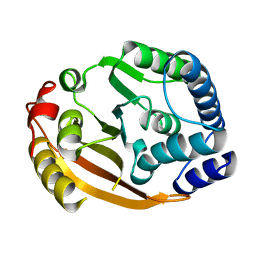 | | Crystal Structure of Gram_Positive Bacillus subtilis Glucose inhibited Division protein B (gidB), Structural genomics, MCSG | | Descriptor: | Methyltransferase gidB | | Authors: | Zhang, R, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-08 | | Release date: | 2004-10-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6A crystal ctructure of Gram-positive Bacillus subtilis glucose inhibited division protein B (gidB)
To be Published
|
|
2O30
 
 | | Nuclear movement protein from E. cuniculi GB-M1 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, NUCLEAR MOVEMENT PROTEIN | | Authors: | Binkowski, T.A, Skarina, T, Onopriyenko, O, Savchenko, A, Edwards, A, Joachimiak, A, MCSG, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-11-30 | | Release date: | 2007-01-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Nuclear movement protein from E. cuniculi GB-M1
TO BE PUBLISHED
|
|
1YXY
 
 | | Crystal Structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes (APC29713) Structural genomics, MCSG | | Descriptor: | Putative N-acetylmannosamine-6-phosphate 2-epimerase | | Authors: | Rotella, F.J, Zhang, R.G, Lezondra, L.E.O, Collart, F.R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6 A crystal structure of putative N-acetylmannosamine-6-P epimerase from Streptococcus pyogenes
To be Published
|
|
2M2J
 
 | | Solution NMR structure of the N-terminal domain of STM1478 from Salmonella typhimurium LT2: Target STR147A of the Northeast Structural Genomics consortium (NESG), and APC101565 of the Midwest Center for Structural Genomics (MCSG). | | Descriptor: | Putative periplasmic protein | | Authors: | Houliston, S, Yee, A, Lemak, A, Garcia, M, Wu, B, Savchenko, A, Montelione, G.T, Arrowsmith, C, Northeast Structural Genomics Consortium (NESG), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-21 | | Release date: | 2013-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
4HYL
 
 | | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365 | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Stage II sporulation protein | | Authors: | Tan, K, Chhor, G, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-13 | | Release date: | 2012-11-28 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | The crystal structure of an anti-sigma-factor antagonist from Haliangium ochraceum DSM 14365
To be Published
|
|
4I66
 
 | |
4I6Z
 
 | | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide | | Descriptor: | DNA OLIGONUCLEOTIDE, Transcriptional regulator, TetR family | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the transcriptional regulator TM1030 with 24bp DNA oligonucleotide
To be Published
|
|
4I4J
 
 | | The structure of SgcE10, the ACP-polyene thioesterase involved in C-1027 biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, ACP-polyene thioesterase, D(-)-TARTARIC ACID, ... | | Authors: | Kim, Y, Bigelow, L, Bearden, J, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.784 Å) | | Cite: | Crystal Structure of Thioesterase SgcE10 Supporting Common Polyene Intermediates in 9- and 10-Membered Enediyne Core Biosynthesis.
Acs Omega, 2, 2017
|
|
4I19
 
 | | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | ACETATE ION, Epoxide hydrolase, FORMIC ACID | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-20 | | Release date: | 2012-12-05 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | The crystal structure of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
4I76
 
 | | Crystal structure of transcriptional regulator TM1030 with octanol | | Descriptor: | 1,2-ETHANEDIOL, OCTAN-1-OL, Transcriptional regulator, ... | | Authors: | Koclega, K.D, Chruszcz, M, Cooper, D.R, Petkowski, J.J, Tkaczuk, K.L, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-11-30 | | Release date: | 2013-01-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of transcriptional regulator TM1030 with octanol
To be Published
|
|
4IC1
 
 | | Crystal structure of SSO0001 | | Descriptor: | IRON/SULFUR CLUSTER, MANGANESE (II) ION, Uncharacterized protein | | Authors: | Nocek, B, Skarina, T, Lemak, S, Beloglazova, N, Flick, R, Brown, G, Savchenko, A, Joachimiak, A, Yakunin, A.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-12-09 | | Release date: | 2013-01-16 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Toroidal structure and DNA cleavage by the CRISPR-associated [4Fe-4S] cluster containing Cas4 nuclease SSO0001 from Sulfolobus solfataricus.
J.Am.Chem.Soc., 135, 2013
|
|
4J10
 
 | | The crystal structure of a secreted protein ESXB (SeMet-labeled) from Bacillus anthracis str. Sterne | | Descriptor: | FORMIC ACID, SECRETED PROTEIN ESXB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-31 | | Release date: | 2013-02-13 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The Crystal Structure of a Secreted Protein Esxb (Semet-Labeled) from Bacillus Anthracis Str. Sterne
To be Published
|
|
4IUS
 
 | | GCN5-related N-acetyltransferase from Kribbella flavida. | | Descriptor: | 1,2-ETHANEDIOL, GCN5-related N-acetyltransferase, MALONATE ION, ... | | Authors: | Osipiuk, J, Chhor, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-01-21 | | Release date: | 2013-01-30 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | GCN5-related N-acetyltransferase from Kribbella flavida.
To be Published
|
|
4J41
 
 | | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-06 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.522 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant P67A) from Bacillus anthracis str. Sterne
To be Published
|
|
4J7J
 
 | | The crystal structure of a secreted protein EsxB (Mutant G53A) from Bacillus anthracis str. Sterne | | Descriptor: | GLYCEROL, secreted protein EsxB | | Authors: | Fan, Y, Tan, K, Chhor, G, Jedrzejczak, R, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-13 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.458 Å) | | Cite: | The crystal structure of a secreted protein EsxB (Mutant, G53A) from Bacillus anthracis str. Sterne
To be Published
|
|
4JDU
 
 | | The crystal structure of an aerotolerance-related membrane protein from Bacteroides fragilis NCTC 9343 with multiple mutations to serines. | | Descriptor: | Aerotolerance-related membrane protein, CALCIUM ION, FORMIC ACID | | Authors: | Fan, Y, Tan, K, Bigelow, L, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-02-25 | | Release date: | 2013-03-27 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The crystal structure of an aerotolerance-related membrane protein from Bacteroides fragilis NCTC 9343 with multiple mutations to serines.
To be Published
|
|
4JOQ
 
 | | Putative ribose ABC transporter, periplasmic solute-binding protein from Rhodobacter sphaeroides | | Descriptor: | 1,2-ETHANEDIOL, ABC ribose transporter, periplasmic solute-binding protein, ... | | Authors: | Osipiuk, J, Tesar, C, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-18 | | Release date: | 2013-04-10 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Putative ribose ABC transporter, periplasmic solute-binding protein from Rhodobacter sphaeroides.
To be Published
|
|
4JGR
 
 | | The crystal structure of sporulation kinase D mutant sensor domain, R131A, from Bacillus subtilis subsp at 2.4A resolution | | Descriptor: | ACETIC ACID, GLYCEROL, Sporulation kinase D | | Authors: | Wu, R, Schiffer, M, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-03-01 | | Release date: | 2013-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insight into the sporulation phosphorelay: Crystal structure of the sensor domain of Bacillus subtilis histidine kinase, KinD.
Protein Sci., 22, 2013
|
|
4KJM
 
 | | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB | | Descriptor: | ACETATE ION, CHLORIDE ION, Extracellular matrix-binding protein ebh, ... | | Authors: | Cymborowski, M, Shabalin, I.G, Joachimiak, G, Chruszcz, M, Gornicki, P, Zhang, R, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-03 | | Release date: | 2013-05-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the Staphylococcus aureus protein (NP_646141.1, domain 3912-4037) similar to streptococcal adhesins emb and ebhA/ebhB
To be Published
|
|
