7W6U
 
 | | Structure of SARS-CoV-2 spike receptor-binding domain complexed with its receptor equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2021-12-02 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses
Nat Commun, 13, 2022
|
|
7XBY
 
 | | The crystal structure of SARS-CoV-2 Omicron BA.1 variant RBD in complex with equine ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, BROMIDE ION, ... | | Authors: | Xu, Z.P, Liu, K.F, Han, P, Qi, J.X. | | Deposit date: | 2022-03-22 | | Release date: | 2022-06-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Binding and structural basis of equine ACE2 to RBDs from SARS-CoV, SARS-CoV-2 and related coronaviruses.
Nat Commun, 13, 2022
|
|
6PH1
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPOL | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.632 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PGZ
 
 | | MTSL labelled T4 lysozyme pseudo-wild type V75C mutant | | Descriptor: | CHLORIDE ION, Endolysin, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PH0
 
 | | T4 lysozyme pseudo-wild type soaked in TEMPO | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin | | Authors: | Cuneo, M.J, Myles, D.A, Li, L. | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.947 Å) | | Cite: | Making hydrogens stand out: Enhanced neutron diffraction from biological crystals using dynamic nuclear polarization
To be published
|
|
6PGY
 
 | |
6LP3
 
 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
7EGS
 
 | | The crystal structure of lobe domain of E. coli RNA polymerase complexed with the C-terminal domain of UvrD | | Descriptor: | DNA helicase II, DNA-directed RNA polymerase subunit beta, GLYCEROL | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
7EGT
 
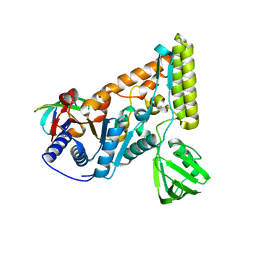 | | The crystal structure of the C-terminal domain of T. thermophilus UvrD complexed with the N-terminal domain of UvrB | | Descriptor: | DNA helicase UvrD, UvrABC system protein B | | Authors: | Zheng, F, Shen, L, Li, L, Zhang, Y. | | Deposit date: | 2021-03-26 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.581 Å) | | Cite: | Crucial role and mechanism of transcription-coupled DNA repair in bacteria.
Nature, 604, 2022
|
|
7DN8
 
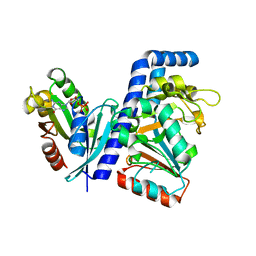 | | Crystal structure of Salmonella effector SopF in complex with ARF1 | | Descriptor: | ADP-ribosylation factor 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ding, J, Shao, F. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6084 Å) | | Cite: | ARF GTPases activate Salmonella effector SopF to ADP-ribosylate host V-ATPase and inhibit endomembrane damage-induced autophagy.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7DN9
 
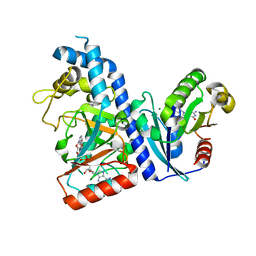 | |
7E7E
 
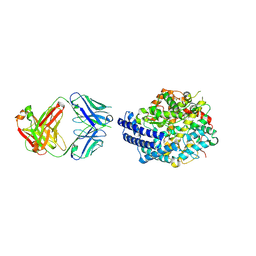 | | The co-crystal structure of ACE2 with Fab | | Descriptor: | Processed angiotensin-converting enzyme 2, ZINC ION, h11B11-Fab | | Authors: | Xiao, J.Y, Zhang, Y. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | A broadly neutralizing humanized ACE2-targeting antibody against SARS-CoV-2 variants.
Nat Commun, 12, 2021
|
|
7EFT
 
 | |
7F7E
 
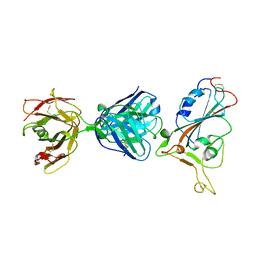 | | SARS-CoV-2 S protein RBD in complex with A5-10 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A5-10 Fab, Light chain of A5-10 Fab, ... | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Etesevimab in combination with JS026 neutralizing SARS-CoV-2 and its variants.
Emerg Microbes Infect, 11, 2022
|
|
3N44
 
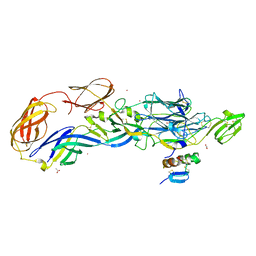 | | Crystal structure of the mature envelope glycoprotein complex (trypsin cleavage; Osmium soak) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N41
 
 | | Crystal structure of the mature envelope glycoprotein complex (spontaneous cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N43
 
 | | Crystal structures of the mature envelope glycoprotein complex (trypsin cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N40
 
 | | Crystal structure of the immature envelope glycoprotein complex of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
3N42
 
 | | Crystal structures of the mature envelope glycoprotein complex (furin cleavage) of Chikungunya virus. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, E1 envelope glycoprotein, ... | | Authors: | Voss, J, Vaney, M.C, Duquerroy, S, Rey, F.A. | | Deposit date: | 2010-05-21 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography.
Nature, 468, 2010
|
|
6ANJ
 
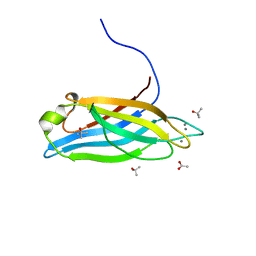 | | Synaptotagmin-7, C2A domain | | Descriptor: | ACETATE ION, CALCIUM ION, Synaptotagmin-7, ... | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6ANK
 
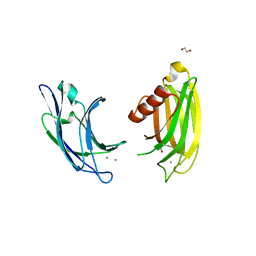 | | Synaptotagmin-7, C2A- and C2B-domains | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Synaptotagmin-7 | | Authors: | Tomchick, D.R, Rizo, J, Voleti, R. | | Deposit date: | 2017-08-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Exceptionally tight membrane-binding may explain the key role of the synaptotagmin-7 C2A domain in asynchronous neurotransmitter release.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
