8CM9
 
 | | Structure of human O-GlcNAc transferase in complex with UDP and tP11 | | Descriptor: | PHE-MET-PRO-LYS-TYR-SER-ILE, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit, URIDINE-5'-DIPHOSPHATE | | Authors: | Meek, R.W, Davies, G.J. | | Deposit date: | 2023-02-18 | | Release date: | 2023-10-04 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phage display uncovers a sequence motif that drives polypeptide binding to a conserved regulatory exosite of O-GlcNAc transferase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4HL8
 
 | | Re-refinement of the vault ribonucleoprotein particle | | Descriptor: | Major vault protein | | Authors: | Casanas, A, Querol-Audi, J, Guerra, P, Pous, J, Tanaka, H, Tsukihara, T, Verdaguer, V, Fita, I. | | Deposit date: | 2012-10-16 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5N8X
 
 | |
5N95
 
 | |
5YT4
 
 | | Galectin-10 variant H53A soaked in glycerol for 5 minutes | | Descriptor: | GLYCEROL, Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Galectin-10: a new structural type of prototype galectin dimer and effects on saccharide ligand binding.
Glycobiology, 28, 2018
|
|
6JK6
 
 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitotriosidase-1 | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-02-27 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6JKF
 
 | | Crystal structure of Serratia marcescens Chitinase B complexed with compound 2-8-s2 | | Descriptor: | 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | Authors: | Yang, Q, Jiang, X. | | Deposit date: | 2019-02-28 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6A62
 
 | | Placental protein 13/galectin-13 variant R53HH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
5DOK
 
 | | Crystal structure of Tetrahymena p45C | | Descriptor: | MAGNESIUM ION, Telomerase associated protein p45 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
6A1T
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A with lactose | | Descriptor: | Galectin-10, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
5E95
 
 | | Crystal Structure of Mb(NS1)/H-Ras Complex | | Descriptor: | GTPase HRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eguchi, R.R, Sha, F, Gupta, A, Koide, A, Koide, S. | | Deposit date: | 2015-10-14 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Inhibition of RAS function through targeting an allosteric regulatory site.
Nat. Chem. Biol., 13, 2017
|
|
6A1U
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33D | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A64
 
 | | Placental protein 13/galectin-13 variant R53HR55NH57RD33G with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A1Y
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant Y35A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6A65
 
 | | Placental protein 13/galectin-13 variant R53HR55N with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.771 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
5ZXL
 
 | | Structure of GldA from E.coli | | Descriptor: | CHLORIDE ION, GLYCEROL, Glycerol dehydrogenase, ... | | Authors: | Zhang, J, Lin, L. | | Deposit date: | 2018-05-21 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Structure of glycerol dehydrogenase (GldA) from Escherichia coli.
Acta Crystallogr F Struct Biol Commun, 75, 2019
|
|
6A1V
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33Q | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
6JMN
 
 | | Crystal structure of Ostrinia furnacalis Chitinase h complexed with compound 2-8-s2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-11-methyl-2-oxidanylidene-7-[[(2R)-oxolan-2-yl]methyl]-N-(pyridin-3-ylmethyl)-1,9-diaza-7-azoniatricyclo[8.4.0.0^{3,8}]tetradeca-3(8),4,6,9,11,13-hexaene-5-carboxamide, Chitinase | | Authors: | Jiang, X, Yang, Q. | | Deposit date: | 2019-03-12 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
J.Med.Chem., 63, 2020
|
|
6A1S
 
 | | Charcot-Leyden crystal protein/Galectin-10 variant E33A | | Descriptor: | Galectin-10 | | Authors: | Su, J. | | Deposit date: | 2018-06-08 | | Release date: | 2018-12-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Identification of key amino acid residues determining ligand binding specificity, homodimerization and cellular distribution of human galectin-10
Glycobiology, 29, 2019
|
|
7DAJ
 
 | | The crystal structure of serotonin N-acetyltransferase in complex with acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAI
 
 | | The crystal structure of a serotonin N-acetyltransferase from Oryza Sativa | | Descriptor: | Serotonin N-acetyltransferase 1, chloroplastic | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6A63
 
 | | Placental protein 13/galectin-13 variant R53HH57R with Lactose | | Descriptor: | Galactoside-binding soluble lectin 13, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
6A66
 
 | | Placental protein 13/galectin-13 variant R53H with Tris | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Galactoside-binding soluble lectin 13 | | Authors: | Su, J.Y. | | Deposit date: | 2018-06-26 | | Release date: | 2018-12-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Resetting the ligand binding site of placental protein 13/galectin-13 recovers its ability to bind lactose
Biosci. Rep., 38, 2018
|
|
7DAL
 
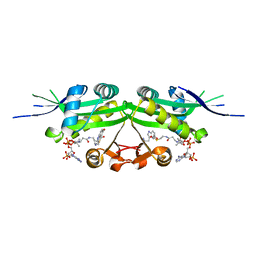 | | The crystal structure of a serotonin N-acetyltransferase in complex with serotonin and acetyl-CoA from Oryza Sativa | | Descriptor: | ACETYL COENZYME *A, SEROTONIN, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7DAK
 
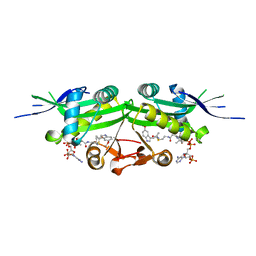 | | The crystal structure of a serotonin N-acetyltransferase in complex with 5-Methoxytryptamine and acetyl-CoA from Oryza Sativa | | Descriptor: | 2-(5-methoxy-1H-indol-3-yl)ethanamine, ACETYL COENZYME *A, Serotonin N-acetyltransferase 1, ... | | Authors: | Zhou, Y.Z, Liao, L.J, Tang, T, Guo, Y, Liu, X.K, Liu, B, Zhao, Y.C. | | Deposit date: | 2020-10-16 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Molecular Dynamics Analysis of Plant Serotonin N-Acetyltransferase Reveal an Acid/Base-Assisted Catalysis in Melatonin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
