3LAM
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-propyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-methyl-5-{[5-(1-methylethyl)-2,6-dioxo-3-propyl-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}benzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
6C7R
 
 | | BRD4 BD1 in complex with compound CF53 | | Descriptor: | Bromodomain-containing protein 4, N-(3-cyclopropyl-1-methyl-1H-pyrazol-5-yl)-7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-9H-pyrimido[4,5-b]indol-4-amine | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
3LAN
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with N1-butyl pyrimidinedione non-nucleoside inhibitor | | Descriptor: | 3-{[3-butyl-5-(1-methylethyl)-2,6-dioxo-1,2,3,6-tetrahydropyrimidin-4-yl]carbonyl}-5-methylbenzonitrile, HIV Reverse transcriptase, SULFATE ION | | Authors: | Lansdon, E.B, Mitchell, M.L. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | N1-Alkyl pyrimidinediones as non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3E9T
 
 | | Crystal structure of Apo-form Calx CBD1 domain | | Descriptor: | CALCIUM ION, Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-09-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of progressive Ca2+ binding states of the Ca2+ sensor Ca2+ binding domain 1 (CBD1) from the CALX Na+/Ca2+ exchanger reveal incremental conformational transitions.
J.Biol.Chem., 285, 2010
|
|
4ZZI
 
 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
5ZHO
 
 | |
4ZZH
 
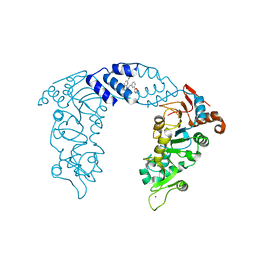 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4ZZJ
 
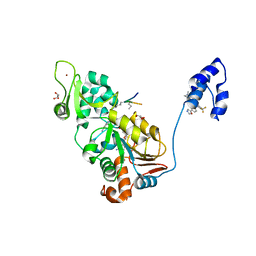 | | SIRT1/Activator/Substrate Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, Ac-p53, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7403 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
7XNA
 
 | | Crystal structure of somatostatin receptor 2 (SSTR2) with peptide antagonist CYN 154806 | | Descriptor: | CYN 154806, Somatostatin receptor type 2,Endo-1,4-beta-xylanase | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
7XN9
 
 | | Crystal structure of SSTR2 and L-054,522 complex | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Somatostatin receptor type 2,Endo-1,4-beta-xylanase, tert-butyl (2S)-6-azanyl-2-[[(2R,3S)-3-(1H-indol-3-yl)-2-[[4-(2-oxidanylidene-3H-benzimidazol-1-yl)piperidin-1-yl]carbonylamino]butanoyl]amino]hexanoate | | Authors: | Zhao, W, Han, S, Qiu, N, Feng, W, Lu, M, Yang, D, Wang, M.-W, Wu, B, Zhao, Q. | | Deposit date: | 2022-04-28 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into ligand recognition and selectivity of somatostatin receptors.
Cell Res., 32, 2022
|
|
6C7Q
 
 | | BRD4 BD2 in complex with compound CE277 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-N-(1-methyl-1H-indazol-3-yl)-9H-pyrimido[4,5-b]indol-4-amine, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2018-01-23 | | Release date: | 2018-08-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-Based Discovery of CF53 as a Potent and Orally Bioavailable Bromodomain and Extra-Terminal (BET) Bromodomain Inhibitor.
J. Med. Chem., 61, 2018
|
|
5ZHG
 
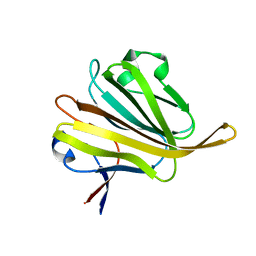 | |
5BPE
 
 | | Crystal structure of EV71 3Cpro in complex with a potent and selective Inhibitor | | Descriptor: | (2~{S})-~{N}-[(1~{R},2~{S})-1-cyano-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepiperidin-3-yl]propan-2-yl]-3-phenyl-2-[[(~{E})-3-phenylprop-2-enoyl]amino]propanamide, EV71 3Cpro | | Authors: | Luqing, S, Yin, Z. | | Deposit date: | 2015-05-28 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Cyanohydrin as an Anchoring Group for Potent and Selective Inhibitors of Enterovirus 71 3C Protease
J.Med.Chem., 58, 2015
|
|
7V4Y
 
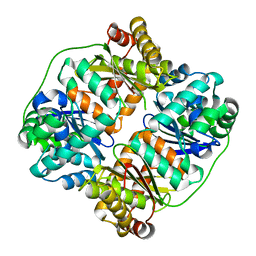 | | TTHA1264/TTHA1265 complex | | Descriptor: | Putative zinc protease, ZINC ION, Zinc-dependent peptidase | | Authors: | Xu, M, Xu, Q, Ran, T, Wang, W, Sun, B, Wang, Q. | | Deposit date: | 2021-08-16 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of TTHA1265 and TTHA1264/TTHA1265 complex reveal an intrinsic heterodimeric assembly.
Int.J.Biol.Macromol., 207, 2022
|
|
3E9U
 
 | | Crystal structure of Calx CBD2 domain | | Descriptor: | Na/Ca exchange protein | | Authors: | Wu, M, Zheng, L. | | Deposit date: | 2008-08-23 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CBD2 from the Drosophila Na(+)/Ca(2+) exchanger: diversity of Ca(2+) regulation and its alternative splicing modification.
J.Mol.Biol., 387, 2009
|
|
6NJS
 
 | | Stat3 Core in complex with compound SD36 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-01-04 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
8T9H
 
 | |
6NUQ
 
 | | Stat3 Core in complex with compound SI109 | | Descriptor: | Signal transducer and activator of transcription 3, [(2-{[(5S,8S,10aR)-3-acetyl-8-({(2S)-5-amino-1-[(diphenylmethyl)amino]-1,5-dioxopentan-2-yl}carbamoyl)-6-oxodecahydropyrrolo[1,2-a][1,5]diazocin-5-yl]carbamoyl}-1H-indol-5-yl)(difluoro)methyl]phosphonic acid (non-preferred name) | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2019-02-01 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo.
Cancer Cell, 36, 2019
|
|
6OJF
 
 | | Dimeric structure of LRRK2 GTPase domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION | | Authors: | Hoang, Q.Q, Wu, C.X, Liao, J, Park, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conformational plasticity in the GTPase domain of the Parkinson's disease-associated protein LRRK2
To be published
|
|
6OJE
 
 | | Dimeric structure of LRRK2 GTPase domain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Leucine-rich repeat serine/threonine-protein kinase 2, MAGNESIUM ION | | Authors: | Hoang, Q.Q, Wu, C.X, Liao, J, Park, Y. | | Deposit date: | 2019-04-11 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for conformational plasticity in the GTPase domain of the Parkinson's disease-associated protein LRRK2
To Be Published
|
|
8X6B
 
 | | Crystal structure of immune receptor PVRIG in complex with ligand Nectin-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nectin-2, Transmembrane protein PVRIG | | Authors: | Hu, S.T, Han, P, Wang, H, Qi, J.X. | | Deposit date: | 2023-11-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the immune recognition and selectivity of the immune receptor PVRIG for ligand Nectin-2.
Structure, 32, 2024
|
|
8YZS
 
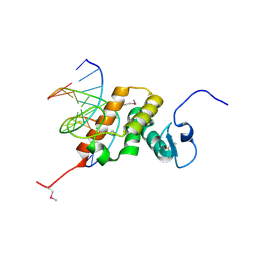 | | Structure of the NACC1 BEN domain in complex with its target DNA | | Descriptor: | CATG-containing DNA, Nucleus accumbens-associated protein 1 | | Authors: | Ren, J, Wang, Z. | | Deposit date: | 2024-04-08 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis of DNA recognition by BEN domain proteins reveals a role for oligomerization in unmethylated DNA selection by BANP.
Nucleic Acids Res., 52, 2024
|
|
8YZT
 
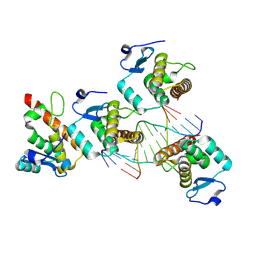 | |
6KUR
 
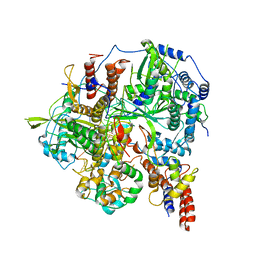 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUK
 
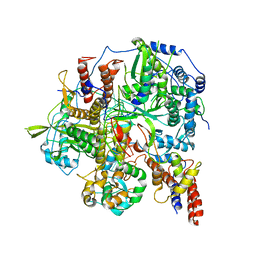 | | Structure of influenza D virus polymerase bound to vRNA promoter in mode A conformation (class A1) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
