3BJE
 
 | |
3BBW
 
 | |
3BBT
 
 | | crystal structure of the ErbB4 kinase in complex with lapatinib | | Descriptor: | N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Qiu, C. | | Deposit date: | 2007-11-11 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
3BZ3
 
 | |
5C25
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 6-((4-((4-cyanophenyl)amino)-1,3,5-triazin-2-yl)amino)-5,7-dimethyl-2-naphthonitrile (JLJ639), a Non-nucleoside Inhibitor | | Descriptor: | 6-({4-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}amino)-5,7- dimethyl-2-naphthonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Chan, A.H, Frey, K.M, Anderson, K.S. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5C24
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 7-((4-((4-cyanophenyl)amino)-1,3,5-triazin-2-yl)amino)-6,8-dimethylindolizine-2-carbonitrile (JLJ605), a non-nucleoside inhibitor | | Descriptor: | 6-({4-[(4-cyanophenyl)amino]-1,3,5-triazin-2-yl}amino)-5,7-dimethylindolizine-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2015-06-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
5AOS
 
 | | Structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A solved at the As edge | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5AOT
 
 | | Very high resolution structure of a novel carbohydrate binding module from Ruminococcus flavefaciens FD-1 endoglucanase Cel5A | | Descriptor: | CACODYLATE ION, Carbohydrate binding module, GLYCEROL | | Authors: | Pires, A.J, Ribeiro, T, Thompson, A, Venditto, I, Fernandes, V.O, Bule, P, Santos, H, Alves, V.D, Pires, V, Ferreira, L.M.A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-09-11 | | Release date: | 2016-06-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5C42
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (K101P) Variant in Complex with 8-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)indolizine-2-carbonitrile (JLJ555), a non-nucleoside inhibitor | | Descriptor: | 8-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}indolizine-2-carbonitrile, HIV-1 Reverse Transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Gray, W.T, Anderson, K.S. | | Deposit date: | 2015-06-17 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Potent Inhibitors Active against HIV Reverse Transcriptase with K101P, a Mutation Conferring Rilpivirine Resistance.
Acs Med.Chem.Lett., 6, 2015
|
|
4MX9
 
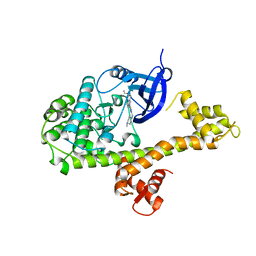 | | CDPK1 from Neospora caninum in complex with inhibitor UW1294 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-[(1-methylpiperidin-4-yl)methyl]-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
4MXA
 
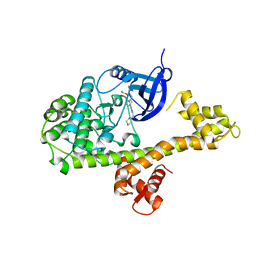 | | CDPK1 from Neospora caninum in complex with inhibitor RM-1-132 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(piperidin-4-ylmethyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Calmodulin-like domain protein kinase isoenzyme gamma, related | | Authors: | Merritt, E.A. | | Deposit date: | 2013-09-26 | | Release date: | 2013-10-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Neospora caninum Calcium-Dependent Protein Kinase 1 Is an Effective Drug Target for Neosporosis Therapy.
Plos One, 9, 2014
|
|
2FVC
 
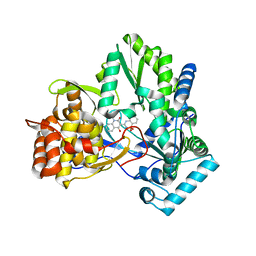 | | Crystal structure of NS5B BK strain (delta 24) in complex with a 3-(1,1-Dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinone | | Descriptor: | 3-(1,1-dioxido-4H-1,2,4-benzothiadiazin-3-yl)-4-hydroxy-1-(3-methylbutyl)quinolin-2(1H)-one, polyprotein | | Authors: | Concha, N.O, Wonacott, A, Singh, O. | | Deposit date: | 2006-01-30 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 3-(1,1-dioxo-2H-(1,2,4)-benzothiadiazin-3-yl)-4-hydroxy-2(1H)-quinolinones, potent inhibitors of hepatitis C virus RNA-dependent RNA polymerase.
J.Med.Chem., 49, 2006
|
|
2JNR
 
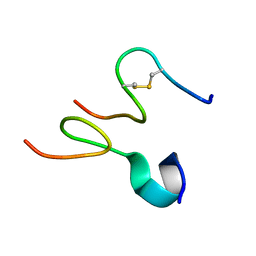 | | Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide | | Descriptor: | ENV polyprotein, VIR165 | | Authors: | Munch, J, Standker, L, Adermann, K, Schulz, A, Pohlmann, S, Chaipan, C, Biet, T, Peters, T, Meyer, B, Wilhelm, D, Lu, H, Jing, W, Jiang, S, Forssmann, W, Kirchhoff, F. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Discovery and Optimization of a Natural HIV-1 Entry Inhibitor Targeting the gp41 Fusion Peptide.
Cell(Cambridge,Mass.), 129, 2007
|
|
2GZQ
 
 | |
1A45
 
 | | GAMMAF CRYSTALLIN FROM BOVINE LENS | | Descriptor: | GAMMAF CRYSTALLIN | | Authors: | Norledge, B.V, Hay, R, Bateman, O.A, Slingsby, C, White, H.E, Moss, D.S, Lindley, P.F, Driessen, H.P.C. | | Deposit date: | 1998-02-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Towards a molecular understanding of phase separation in the lens: a comparison of the X-ray structures of two high Tc gamma-crystallins, gammaE and gammaF, with two low Tc gamma-crystallins, gammaB and gammaD.
Exp.Eye Res., 65, 1997
|
|
7TV2
 
 | | X-ray crystal structure of HIV-2 CA protein CTD | | Descriptor: | Capsid protein p24 | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2022-02-03 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HIV-2 Immature Particle Morphology Provides Insights into Gag Lattice Stability and Virus Maturation.
J.Mol.Biol., 435, 2023
|
|
7UXO
 
 | | Structure of PDL1 in complex with FP30790, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP30790, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UYK
 
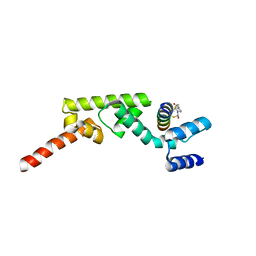 | | Structure of RNF31 in complex with FP06655, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06655, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UWI
 
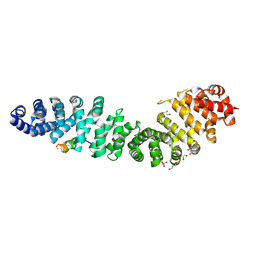 | | Structure of beta-catenin in complex with FP01567, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, GLYCEROL, Helicon Polypeptide FP01567, ... | | Authors: | Brennan, M, Agarwal, S, Thomson, T, Wahl, S, Ramirez, J, Verdine, G, McGee, J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXQ
 
 | | Structure of PDL1 in complex with FP28135, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, FP28135, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UWO
 
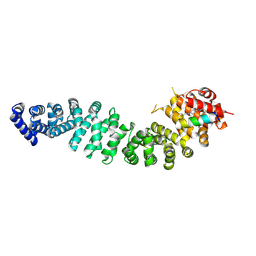 | | Structure of beta-catenin in complex with FP05874, a Helicon Polypeptide | | Descriptor: | Catenin beta-1, Helicon Polypeptide FP05874, N,N'-(1,4-phenylene)diacetamide | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Ramirez, J, Hriniak, B, Verdine, G, McGee, J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXK
 
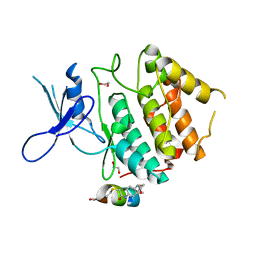 | | Structure of CDK2 in complex with FP24322, a Helicon Polypeptide | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, FP24322, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXP
 
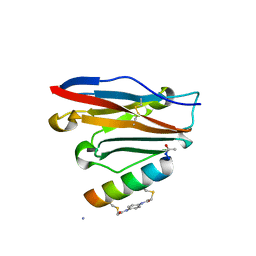 | | Structure of PDL1 in complex with FP28132, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, FP28132, N,N'-(1,4-phenylene)diacetamide, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Travaline, T, Tattersfield, H, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXI
 
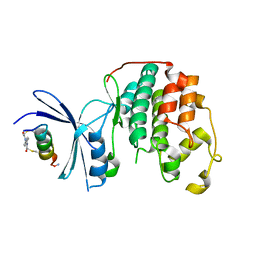 | | Structure of CDK2 in complex with FP19711, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, Cyclin-dependent kinase 2, FP19711, ... | | Authors: | Li, K, Agarwal, S, Tokareva, O, Thomson, T, Wahl, S, Verdine, G, McGee, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UY2
 
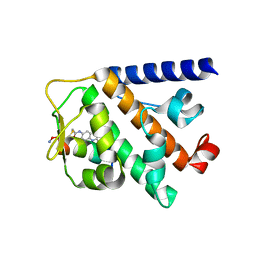 | | Structure of RNF31 in complex with FP06649, a Helicon Polypeptide | | Descriptor: | AMINO GROUP, E3 ubiquitin-protein ligase RNF31, Helicon FP06649, ... | | Authors: | Agarwal, S, Thomson, T, Wahl, S, Walkup, W, Olsen, T, Verdine, G, McGee, J. | | Deposit date: | 2022-05-06 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | De novo mapping of alpha-helix recognition sites on protein surfaces using unbiased libraries.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
