1OMU
 
 | | SOLUTION STRUCTURE OF OVOMUCOID (THIRD DOMAIN) FROM DOMESTIC TURKEY (298K, PH 4.1) (NMR, 50 STRUCTURES) (REFINED MODEL USING NETWORK EDITING ANALYSIS) | | Descriptor: | OVOMUCOID (THIRD DOMAIN) | | Authors: | Hoogstraten, C.G, Choe, S, Westler, W.M, Markley, J.L. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Comparison of the accuracy of protein solution structures derived from conventional and network-edited NOESY data.
Protein Sci., 4, 1995
|
|
7N8U
 
 | |
5HL4
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | COBALT HEXAMMINE(III), FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-14 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
1WP5
 
 | |
5HQD
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CALCIUM ION, Thermolysin, ZINC ION | | Authors: | Roesser, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2016-01-21 | | Release date: | 2016-02-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
3VEJ
 
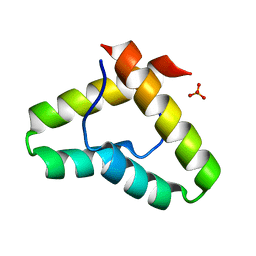 | | Crystal structure of the Get5 carboxyl domain from S. cerevisiae | | Descriptor: | PHOSPHATE ION, Ubiquitin-like protein MDY2 | | Authors: | Chartron, J.W, Vandervelde, D.G, Rao, M, Clemons Jr, W.M. | | Deposit date: | 2012-01-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Get5 Carboxyl-terminal Domain Is a Novel Dimerization Motif That Tethers an Extended Get4/Get5 Complex.
J.Biol.Chem., 287, 2012
|
|
1XRK
 
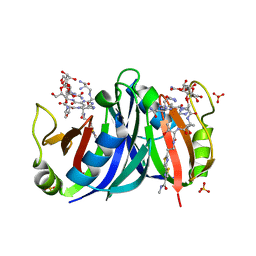 | | Crystal structure of a mutant bleomycin binding protein from Streptoalloteichus hindustanus displaying increased thermostability | | Descriptor: | BLEOMYCIN A2, Bleomycin resistance protein, SULFATE ION | | Authors: | Brouns, S.J.J, Wu, H, Akerboom, J, Turnbull, A.P, de Vos, W.M, Van der Oost, J. | | Deposit date: | 2004-10-15 | | Release date: | 2005-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering a selectable marker for hyperthermophiles
J.Biol.Chem., 280, 2005
|
|
1XOE
 
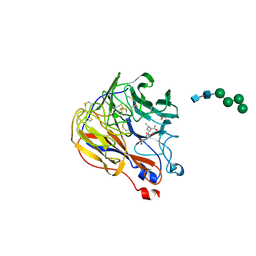 | | N9 Tern influenza neuraminidase complexed with (2R,4R,5R)-5-(1-Acetylamino-3-methyl-butyl-pyrrolidine-2, 4-dicarobyxylic acid 4-methyl esterdase complexed with | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-4-(METHOXYCARBONYL)PROLINE, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase
containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
6OVU
 
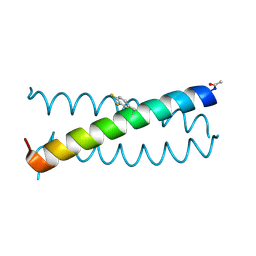 | | Coiled-coil Trimer with Glu:3,4-difluorophenylalanine:Lys Triad | | Descriptor: | Coiled-coil Trimer with Glu:3,4-difluorophenylalanine:Lys Triad | | Authors: | Smith, M.S, Stern, K.L, Billings, W.M, Price, J.L. | | Deposit date: | 2019-05-08 | | Release date: | 2020-04-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Context-Dependent Stabilizing Interactions among Solvent-Exposed Residues along the Surface of a Trimeric Helix Bundle.
Biochemistry, 59, 2020
|
|
3SZ7
 
 | |
1XOG
 
 | | N9 Tern Influenza neuraminidase complexed with a 2,5-Disubstituted tetrahydrofuran-5-carboxylic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-2,5-ANHYDRO-3,4-DIDEOXY-4-(METHOXYCARBONYL)PENTONIC ACID, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1XS8
 
 | |
3UG7
 
 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
3UG6
 
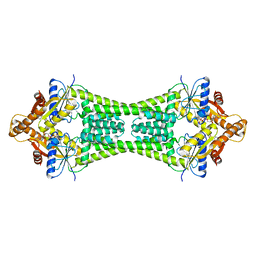 | | Crystal Structure of Get3 from Methanocaldococcus jannaschii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Suloway, C.J.M, Rome, M.E, Clemons Jr, W.M. | | Deposit date: | 2011-11-02 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Tail-anchor targeting by a Get3 tetramer: the structure of an archaeal homologue.
Embo J., 31, 2012
|
|
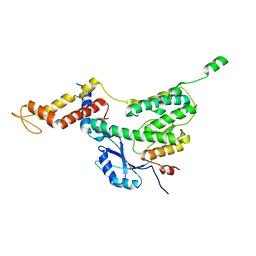
6F7J
 
 | |
1A32
 
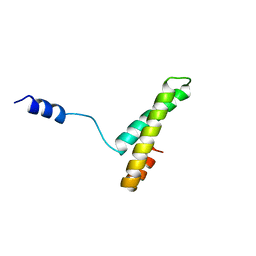 | |
8TLU
 
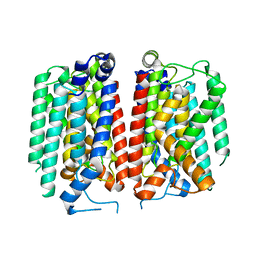 | | E. coli MraY mutant-T23P | | Descriptor: | Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Orta, A.K, Li, Y.E, Clemons, W.M. | | Deposit date: | 2023-07-27 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Synthesis of lipid-linked precursors of the bacterial cell wall is governed by a feedback control mechanism in Pseudomonas aeruginosa.
Nat Microbiol, 9, 2024
|
|
8QAC
 
 | | X-ray crystal structure of a de novo designed antiparallel coiled-coil 8-helix bundle with 4 heptad repeats, antiparallel 8-helix bundle-GLIA | | Descriptor: | antiparallel 8-helix bundle-GLIA | | Authors: | Albanese, K.I, Dawson, W.M, Petrenas, R, Woolfson, D.N. | | Deposit date: | 2023-08-22 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rationally seeded computational protein design of ɑ-helical barrels.
Nat.Chem.Biol., 20, 2024
|
|
7KWZ
 
 | | TDP-43 LCD amyloid fibrils | | Descriptor: | Isoform 2 of TAR DNA-binding protein 43 | | Authors: | Li, Q, Babinchak, W.M, Surewicz, W.K. | | Deposit date: | 2020-12-02 | | Release date: | 2021-02-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Nat Commun, 12, 2021
|
|
1B3B
 
 | | THERMOTOGA MARITIMA GLUTAMATE DEHYDROGENASE MUTANT N97D, G376K | | Descriptor: | PROTEIN (GLUTAMATE DEHYDROGENASE) | | Authors: | Knapp, S, Lebbink, J.H.G, Van Der Oost, J, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-07 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering activity and stability of Thermotoga maritima glutamate dehydrogenase. I. Introduction of a six-residue ion-pair network in the hinge region.
J.Mol.Biol., 280, 1998
|
|
1B26
 
 | | GLUTAMATE DEHYDROGENASE | | Descriptor: | GLUTAMATE DEHYDROGENASE | | Authors: | Knapp, S, Devos, W.M, Rice, D, Ladenstein, R. | | Deposit date: | 1998-12-04 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of glutamate dehydrogenase from the hyperthermophilic eubacterium Thermotoga maritima at 3.0 A resolution.
J.Mol.Biol., 267, 1997
|
|
7KB0
 
 | | O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima with pyridoxal-5-phosphate (PLP) bound in the internal aldimine state | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
7KB1
 
 | | Complex of O-acety-L-homoserine aminocarboxypropyltransferase (MetY) from Thermotoga maritima and a key reaction intermediate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]but-3-enoic acid, MAGNESIUM ION, O-acetyl-L-homoserine sulfhydrylase, ... | | Authors: | Brewster, J.L, Pachl, P, Squire, C, Selmer, M, Patrick, W.M. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures and kinetics of Thermotoga maritima MetY reveal new insights into the predominant sulfurylation enzyme of bacterial methionine biosynthesis.
J.Biol.Chem., 296, 2021
|
|
4WR3
 
 | | Y274F alanine racemase from E. coli | | Descriptor: | Alanine racemase, biosynthetic, GLYCEROL, ... | | Authors: | Squire, C.J, Yosaatmadja, Y, Patrick, W.M. | | Deposit date: | 2014-10-23 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Evolutionary Insights from the Reciprocal Promiscuity of Two Pyridoxal Phosphate-dependent Enzymes.
J.Biol.Chem., 291, 2016
|
|
3NTI
 
 | | Crystal structure of Tudor and Aubergine [R15(me2s)] complex | | Descriptor: | Maternal protein tudor, peptide from Aubergine | | Authors: | Liu, H.P, Huang, Y, Li, Z.Z, Gong, W.M, Xu, R.M. | | Deposit date: | 2010-07-05 | | Release date: | 2010-09-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for methylarginine-dependent recognition of Aubergine by Tudor
Genes Dev., 24, 2010
|
|
