8V31
 
 | |
5WC8
 
 | |
5WC6
 
 | |
2FYV
 
 | | Golgi alpha-mannosidase II complex with an amino-salacinol carboxylate analog | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-DEOXY-6-[(2R,3R,4R)-3,4-DIHYDROXY-2-(HYDROXYMETHYL)PYRROLIDIN-1-YL]-L-GULONIC ACID, ... | | Authors: | Kuntz, D.A, Hamlet, T, Rose, D.R. | | Deposit date: | 2006-02-08 | | Release date: | 2006-12-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis, enzymatic activity, and X-ray crystallography of an unusual class of amino acids.
Bioorg.Med.Chem., 14, 2006
|
|
9ATN
 
 | |
5OMZ
 
 | |
2AEI
 
 | | Crystal structure of a ternary complex of factor VIIa/tissue factor and 2-[[6-[3-(aminoiminomethyl)phenoxy]-3,5-difluro-4-[(1-methyl-3-phenylpropyl)amino]-2-pyridinyl]oxy]-benzoic acid | | Descriptor: | 2-({6-{3-[AMINO(IMINO)METHYL]PHENOXY}-3,5-DIFLUORO-4-[(1-METHYL-3-PHENYLPROPYL)AMINO]-2-PYRIDINYL}OXY)BENZOIC ACID, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Adler, M, Whitlow, M. | | Deposit date: | 2005-07-22 | | Release date: | 2006-08-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The discovery of fluoropyridine-based inhibitors of the Factor VIIa/TF complex.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
8D47
 
 | |
8D48
 
 | | sd1.040 Fab in complex with SARS-CoV-2 Spike 2P glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, sd1.040 Fab heavy chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O. | | Deposit date: | 2022-06-01 | | Release date: | 2023-01-25 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Human neutralizing antibodies to cold linear epitopes and subdomain 1 of the SARS-CoV-2 spike glycoprotein.
Sci Immunol, 8, 2023
|
|
4WTB
 
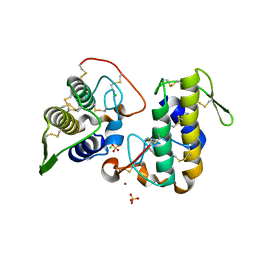 | | BthTX-I, a svPLA2s-like toxin, complexed with zinc ions | | Descriptor: | Basic phospholipase A2 homolog bothropstoxin-1, CHLORIDE ION, SULFATE ION, ... | | Authors: | Borges, R.J, Fontes, M.R.M. | | Deposit date: | 2014-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Functional and structural studies of a Phospholipase A2-like protein complexed to zinc ions: Insights on its myotoxicity and inhibition mechanism.
Biochim. Biophys. Acta, 1861, 2017
|
|
2RHS
 
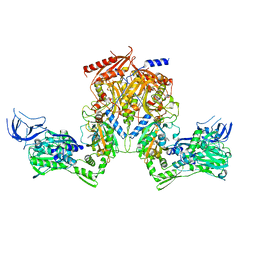 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
2RHQ
 
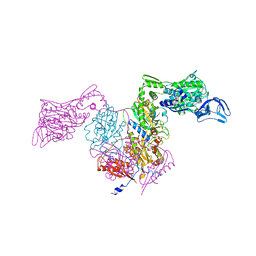 | | PheRS from Staphylococcus haemolyticus- rational protein engineering and inhibitor studies | | Descriptor: | 1-{3-[(4-pyridin-2-ylpiperazin-1-yl)sulfonyl]phenyl}-3-(1,3-thiazol-2-yl)urea, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Evdokimov, A.G, Mekel, M. | | Deposit date: | 2007-10-09 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational protein engineering in action: the first crystal structure of a phenylalanine tRNA synthetase from Staphylococcus haemolyticus.
J.Struct.Biol., 162, 2008
|
|
5Y0A
 
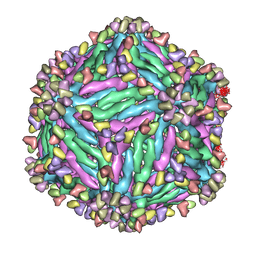 | |
7BN2
 
 | |
7BN3
 
 | |
7BN1
 
 | |
7ZX4
 
 | | Clathrin N-terminal domain in complex with a HURP phospho-peptide | | Descriptor: | CHLORIDE ION, Clathrin heavy chain 1, Disks large-associated protein 5, ... | | Authors: | Kliche, J, Badgujar, D, Dobritzsch, D, Ivarsson, Y. | | Deposit date: | 2022-05-20 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Large-scale phosphomimetic screening identifies phospho-modulated motif-based protein interactions.
Mol.Syst.Biol., 19, 2023
|
|
2V59
 
 | |
2V58
 
 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2V5A
 
 | |
2VD4
 
 | | Structure of small-molecule inhibitor of Glmu from Haemophilus influenzae reveals an allosteric binding site | | Descriptor: | 4-chloro-N-(3-methoxypropyl)-N-[(3S)-1-(2-phenylethyl)piperidin-3-yl]benzamide, BIFUNCTIONAL PROTEIN GLMU, MAGNESIUM ION, ... | | Authors: | Mochalkin, I, Lightle, S, McDowell, L. | | Deposit date: | 2007-09-28 | | Release date: | 2008-01-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a Small-Molecule Inhibitor Complexed with Glmu from Haemophilus Influenzae Reveals an Allosteric Binding Site.
Protein Sci., 17, 2008
|
|
2W0W
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 2 | | Descriptor: | GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, N-{6-(CYCLOPROPYLMETHOXY)-7-METHOXY-2-[6-(2-METHYLPROPYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]QUINAZOLIN-4-YL}-2,2,2-TRIFLUOROETHANESULFONAMIDE, TETRAETHYLENE GLYCOL | | Authors: | Mochalkin, I, Melnick, M. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
2W0V
 
 | | Crystal structure of Glmu from Haemophilus influenzae in complex with quinazoline inhibitor 1 | | Descriptor: | 6-(CYCLOPROP-2-EN-1-YLMETHOXY)-2-[6-(CYCLOPROPYLMETHYL)-5-OXO-3,4,5,6-TETRAHYDRO-2,6-NAPHTHYRIDIN-2(1H)-YL]-7-METHOXYQUINAZOLIN-4(3H)-ONE, GLUCOSAMINE-1-PHOSPHATE N-ACETYLTRANSFERASE, SULFATE ION, ... | | Authors: | Mochalkin, I, Melnick, M. | | Deposit date: | 2008-10-10 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Initial Sar of Quinazoline Inhibitors of Glmu from Haemophilus Influenzae
To be Published
|
|
2F9Y
 
 | | The Crystal Structure of The Carboxyltransferase Subunit of ACC from Escherichia coli | | Descriptor: | Acetyl-CoA carboxylase, Carboxyltransferase alpha chain, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-06 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Structure of the Carboxyltransferase Component of Acetyl-CoA Carboxylase Reveals a Zinc-Binding Motif Unique to the Bacterial Enzyme(,).
Biochemistry, 45, 2006
|
|
2F9I
 
 | | Crystal Structure of the carboxyltransferase subunit of ACC from Staphylococcus aureus | | Descriptor: | ZINC ION, acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, acetyl-coenzyme A carboxylase carboxyl transferase subunit beta | | Authors: | Bilder, P.W. | | Deposit date: | 2005-12-05 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The structure of the carboxyltransferase component of acetyl-coA carboxylase reveals a zinc-binding motif unique to the bacterial enzyme.
Biochemistry, 45, 2006
|
|
