1FIY
 
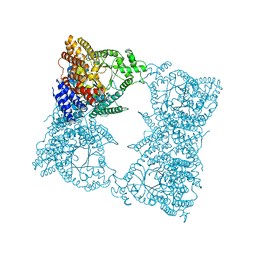 | | THREE-DIMENSIONAL STRUCTURE OF PHOSPHOENOLPYRUVATE CARBOXYLASE FROM ESCHERICHIA COLI AT 2.8 A RESOLUTION | | Descriptor: | ASPARTIC ACID, PHOSPHOENOLPYRUVATE CARBOXYLASE | | Authors: | Kai, Y, Matsumura, H, Inoue, T, Terada, K, Nagara, Y, Yoshinaga, T, Kihara, A, Izui, K. | | Deposit date: | 1998-05-02 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of phosphoenolpyruvate carboxylase: a proposed mechanism for allosteric inhibition.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
5FDG
 
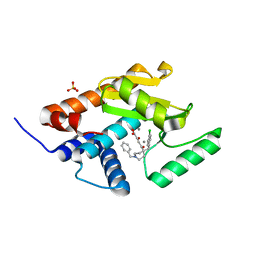 | | Endonuclease inhibitor 3 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2015-12-16 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
5FDD
 
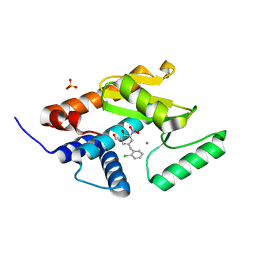 | | Endonuclease inhibitor 1 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2015-12-16 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
5I13
 
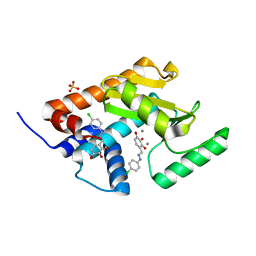 | | Endonuclease inhibitor 2 bound to influenza strain H1N1 polymerase acidic subunit N-terminal region at pH 7.0 | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.151 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
1SIO
 
 | | Structure of Kumamolisin-As complexed with a covalently-bound inhibitor, AcIPF | | Descriptor: | Ace-ILE-PRO-PHL peptide inhibitor, CALCIUM ION, SULFATE ION, ... | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
1SIU
 
 | | KUMAMOLISIN-AS E78H MUTANT | | Descriptor: | CALCIUM ION, SULFATE ION, kumamolisin-As | | Authors: | Li, M, Wlodawer, A, Gustchina, A, Tsuruoka, N, Ashida, M, Minakata, H, Oyama, H, Oda, K, Nishino, T, Nakayama, T. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-As, a serine-carboxyl peptidase with collagenase activity
J.Biol.Chem., 279, 2004
|
|
4ZHZ
 
 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region with expelling one of the metal ions in the active site | | Descriptor: | 5-(2-chlorobenzyl)-2-hydroxy-3-nitrobenzaldehyde, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
6JKB
 
 | | Crystal structure of metallo-beta-lactamse, NDM-1, in complex with hydrolyzed ampicillin | | Descriptor: | (2R,4S)-2-[(R)-{[(2R)-2-amino-2-phenylacetyl]amino}(carboxy)methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
4YYL
 
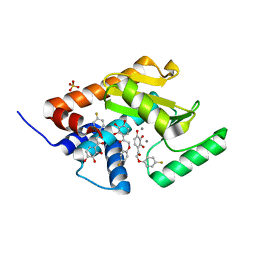 | | Phenolic acid derivative bound to influenza strain H1N1 polymerase subunit PA endonuclease | | Descriptor: | 2-(4-fluorophenoxy)-1-(2,3,4-trihydroxyphenyl)ethanone, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-03-24 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Structural and computational study on inhibitory compounds for endonuclease activity of influenza virus polymerase
Bioorg.Med.Chem., 23, 2015
|
|
4ZI0
 
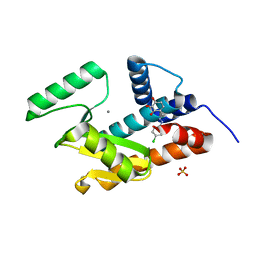 | | Endonuclease inhibitor bound to influenza strain H1N1 polymerase acidic subunit N-terminal region without a chelation to the metal ions in the active site | | Descriptor: | 4-{(E)-[2-(4-chlorophenyl)hydrazinylidene]methyl}benzene-1,2,3-triol, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-04-27 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit
Biochemistry, 55, 2016
|
|
4ZQQ
 
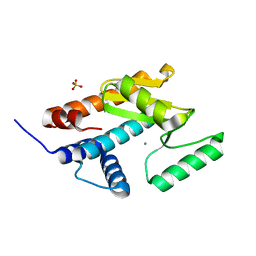 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, SULFATE ION | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Neya, S, Hoshino, T. | | Deposit date: | 2015-05-11 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Distinctive Binding Modes of Endonuclease Inhibitors to the N-Terminal Region of Influenza Virus Polymerase Acidic Subunit.
Biochemistry, 55, 2016
|
|
2Z9S
 
 | | Crystal Structure Analysis of rat HBP23/Peroxiredoxin I, Cys52Ser mutant | | Descriptor: | Peroxiredoxin-1 | | Authors: | Matsumura, T, Okamoto, K, Nishino, T, Abe, Y. | | Deposit date: | 2007-09-25 | | Release date: | 2007-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimer-Oligomer Interconversion of Wild-type and Mutant Rat 2-Cys Peroxiredoxin: DISULFIDE FORMATION AT DIMER-DIMER INTERFACES IS NOT ESSENTIAL FOR DECAMERIZATION
J.Biol.Chem., 283, 2008
|
|
6JKA
 
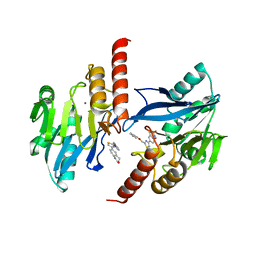 | | Crystal structure of metallo-beta-lactamse, IMP-1, in complex with a thiazole-bearing inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-(phenoxymethyl)-1,3-thiazol-4-yl]phenyl]propanoic acid, 6-[2-(phenoxymethyl)-1,3-thiazol-4-yl]-3,4-dihydro-1H-quinolin-2-one, Metallo-beta-lactamase type 2, ... | | Authors: | Kamo, T, Kuroda, K, Kondo, S, Hayashi, U, Fudo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-28 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Identification of the Inhibitory Compounds for Metallo-beta-lactamases and Structural Analysis of the Binding Modes.
Chem Pharm Bull (Tokyo), 69, 2021
|
|
6JE7
 
 | | Crystal structure of human serum albumin crystallized by ammonium sulfate | | Descriptor: | Serum albumin | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
1SN7
 
 | | KUMAMOLISIN-AS, APOENZYME | | Descriptor: | CALCIUM ION, kumamolisin-As | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Oda, K, Nishino, T. | | Deposit date: | 2004-03-10 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and biochemical investigations of kumamolisin-as, a serine-carboxyl peptidase with collagenase activity.
J.Biol.Chem., 279, 2004
|
|
7XGC
 
 | |
7XGQ
 
 | |
7XJ5
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIS
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | (2-methoxy-4-methoxycarbonyl-phenyl) 5-nitrofuran-2-carboxylate, MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIU
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XIT
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-14 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ7
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, ZINC ION, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
7XJ4
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with nitrofuran methoxy(methoxycarbonyl)phenyl ester | | Descriptor: | MANGANESE (II) ION, Reverse Transcriptase RNase H domain, S-[5-[(E)-2-phenylethenyl]-1,3,4-oxadiazol-2-yl] 5-nitrothiophene-2-carbothioate, ... | | Authors: | Lu, H, Komukai, Y, Usami, K, Guo, Y, Qiao, X, Nukaga, M, Hoshino, T. | | Deposit date: | 2022-04-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computational and Crystallographic Analysis of Binding Structures of Inhibitory Compounds for HIV-1 RNase H Activity.
J.Chem.Inf.Model., 62, 2022
|
|
2E1Q
 
 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
5ZXW
 
 | | Crystal structure of human carbonic anhydrase II crystallized by ammonium sulfate | | Descriptor: | Carbonic anhydrase 2, SULFATE ION, ZINC ION | | Authors: | Kitahara, M, Fudo, S, Yoneda, T, Nukaga, M, Hoshino, T. | | Deposit date: | 2018-05-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.316 Å) | | Cite: | Anisotropic Distribution of Ammonium Sulfate Ions in Protein Crystallization
Cryst.Growth Des., 19, 2019
|
|
