7A5S
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Homogeneous Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A5R
 
 | | Complex of SARS-CoV-2 spike and CR3022 Fab (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab Heavy Chain, CR3022 Fab Light Chain, ... | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-08-21 | | Release date: | 2020-09-16 | | Last modified: | 2020-11-04 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein.
Nat Commun, 11, 2020
|
|
7A95
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound and 1 RBD Erect in Clockwise Direction | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A91
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Non-Uniform Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A92
 
 | | Dissociated S1 domain of SARS-CoV-2 Spike bound to ACE2 (Unmasked Refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A94
 
 | | SARS-CoV-2 Spike Glycoprotein with 1 ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
6ZGG
 
 | |
6ZGE
 
 | | Uncleavable Spike Protein of SARS-CoV-2 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Wrobel, A.G, Benton, D.J, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7A97
 
 | | SARS-CoV-2 Spike Glycoprotein with 2 ACE2 Bound | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Benton, D.J, Wrobel, A.G, Rosenthal, P.B, Gamblin, S.J. | | Deposit date: | 2020-09-01 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature, 588, 2020
|
|
7A93
 
 | |
6J05
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | ARSENIC, SODIUM ION, Transcriptional regulator ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-21 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
6J0E
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARSENIC, Arsenic responsive repressor ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
8R84
 
 | | pentameric IgMFc-AIM complex focused refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CD5 antigen-like, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
8R83
 
 | | pentameric IgMFc-AIM complex global refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q, Arai, S, Miyazaki, T, Rosenthal, P. | | Deposit date: | 2023-11-28 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM reveals structural basis for human AIM/CD5L recognition of polymeric immunoglobulin M.
Nat Commun, 15, 2024
|
|
7QDO
 
 | | Cryo-EM structure of human monomeric IgM-Fc | | Descriptor: | Isoform 2 of Immunoglobulin heavy constant mu | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2021-11-27 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8ADZ
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE3
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE2
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8AE0
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8ADY
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
8BPG
 
 | | FcMR binding at subunit Fcu3 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPF
 
 | | FcMR binding at subunit Fcu1 of IgM pentamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8BPE
 
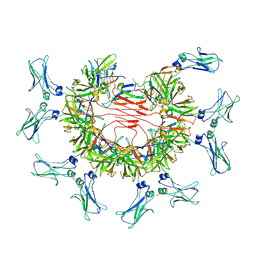 | | 8:1 binding of FcMR on IgM pentameric core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fas apoptotic inhibitory molecule 3, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-11-16 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structural basis for Fc receptor recognition of immunoglobulin M.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7NT9
 
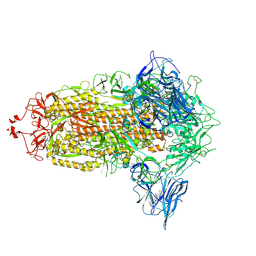 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7NTA
 
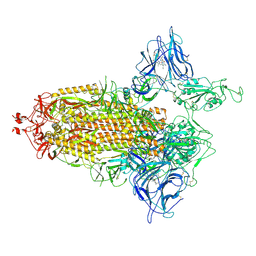 | | Trimeric SARS-CoV-2 spike ectodomain in complex with biliverdin (one RBD erect) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Rosa, A, Pye, V.E, Nans, A, Cherepanov, P. | | Deposit date: | 2021-03-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
