6WVX
 
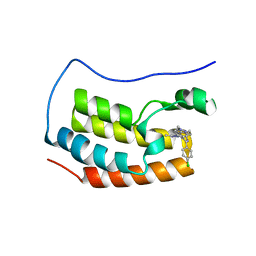 | | Crystal structure of the first bromodomain of human BRD4 with benzodiazepine inhibitor | | Descriptor: | 6'-(4-chlorophenyl)-1'-methyl-8'-(1-methyl-1H-pyrazol-4-yl)spiro[cyclopropane-1,4'-[1,2,4]triazolo[4,3-a][1,4]benzodiazepine], Bromodomain-containing protein 4 | | Authors: | Eron, S.J. | | Deposit date: | 2020-05-07 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural Characterization of Degrader-Induced Ternary Complexes Using Hydrogen-Deuterium Exchange Mass Spectrometry and Computational Modeling: Implications for Structure-Based Design.
Acs Chem.Biol., 16, 2021
|
|
3DXJ
 
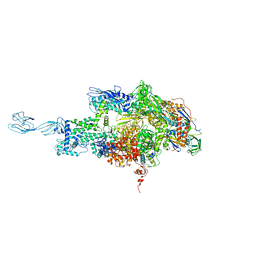 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
4AJJ
 
 | | rat LDHA in complex with 2-((3,4-dimethoxyphenyl)methyl))propanedioic acid and N-(2-methyl-1,3-benzothiazol-6-yl)-3-ureido-propanamide | | Descriptor: | 2-((3,4-DIMETHOXYPHENYL)METHYL))PROPANEDIOIC ACID, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G, Vogtherr, M, Ward, R, Tart, J, Davies, G, Patel, J, Greenwood, R. | | Deposit date: | 2012-02-16 | | Release date: | 2012-03-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Synthesis of Novel Lactate Dehydrogenase a Inhibitors by Fragment-Based Lead Generation
J.Med.Chem., 55, 2012
|
|
6RJZ
 
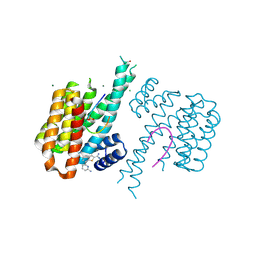 | | Fragment AZ-015 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 7-(6-azanylpyridin-2-yl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-29 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RKI
 
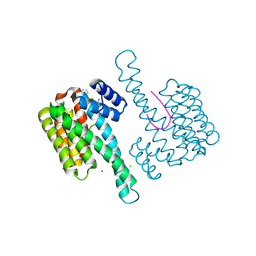 | | Fragment AZ-023 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-[(3-aminophenyl)amino]-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-30 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6S3C
 
 | | Fragment AZ-019 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-phenyl-5-(piperidin-4-ylmethyl)thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6R5L
 
 | | Fragment AZ-006 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 4-[[(2~{S})-1-azanylpropan-2-yl]amino]-6-(sulfanylmethyl)-1-benzothiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-03-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.884 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6S39
 
 | | Fragment AZ-018 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-(3-azanylpropyl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-24 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
2YWP
 
 | | Crystal Structure of CHK1 with a Urea Inhibitor | | Descriptor: | 1-(5-CHLORO-2,4-DIMETHOXYPHENYL)-3-(5-CYANOPYRAZIN-2-YL)UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and biological evaluation of 1-(2,4,5-trisubstituted phenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent Chk1 kinase inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
6RHC
 
 | | Fragment AZ-003 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-azanyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Genet, S, Wolter, M, Guillory, X, Somsen, B, Leysen, S, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-04-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
6RNU
 
 | | BCL-XL in a complex with a covalent small molecule inhibitor | | Descriptor: | 4-(4-fluorophenyl)-3-fluorosulfonyl-benzoic acid, BROMIDE ION, Bcl-2-like protein 1 | | Authors: | Hargreaves, D. | | Deposit date: | 2019-05-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery and optimization of covalent Bcl-xL antagonists.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6SLX
 
 | | Fragment AZ-010 binding at the TAZpS89/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, TAZpS89, ~{N}-[3-(5-carbamimidoylthiophen-3-yl)phenyl]propanamide | | Authors: | Ottmann, C, Wolter, M, Guillory, X, Genet, S, Somsen, B, Leysen, S, Castaldi, P. | | Deposit date: | 2019-08-20 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.800045 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
5TMF
 
 | | Re-refinement of thermus thermophilus RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Wang, J. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | On the validation of crystallographic symmetry and the quality of structures.
Protein Sci., 24, 2015
|
|
6ZCU
 
 | | syk in complex with 57262_SYKB-AZ13344324-2 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 2b
To Be Published
|
|
6ZCR
 
 | | SYK Kinase domain in complex with azabenzimidazole inhibitor 7 | | Descriptor: | 6-chloro-2-(2-fluoro-4,5-dimethoxyphenyl)-N-(piperidin-4-ylmethyl)-3H-imidazo[4,5-b]pyridin-7-amine, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2020-06-12 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | SYK Kinase domain in complex with azabenzimidazole inhibitor 7
To Be Published
|
|
2LQR
 
 | | NMR structure of Ig3 domain of palladin | | Descriptor: | Palladin | | Authors: | Beck, M.R, Dixon IV, R.D.S, Otey, C.A, Campbell, S.L, Murphy, G.S. | | Deposit date: | 2012-03-13 | | Release date: | 2013-01-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Function of Palladin's Actin Binding Domain.
J.Mol.Biol., 425, 2013
|
|
2OAE
 
 | | Crystal structure of rat dipeptidyl peptidase (DPPIV) with thiazole-based peptide mimetic #31 | | Descriptor: | Dipeptidyl peptidase 4, N-{[(3S,5S)-5-(1,3-THIAZOLIDIN-3-YLCARBONYL)PYRROLIDIN-3-YL]METHYL}-1,3-THIAZOLE-4-CARBOXAMIDE, SULFATE ION | | Authors: | Longenecker, K.L, Shuai, Q, Patel, J, Wiedeman, P. | | Deposit date: | 2006-12-15 | | Release date: | 2007-02-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
