4AUA
 
 | | Liganded X-ray crystal structure of cyclin dependent kinase 6 (CDK6) | | Descriptor: | 1H-benzimidazol-2-yl(1H-pyrrol-2-yl)methanone, CYCLIN-DEPENDENT KINASE 6 | | Authors: | Cho, Y.S, Angove, H, Brain, C, Chen, C.H.T, Cheng, R, Chopra, R, Chung, K, Congreve, M, Dagostin, C, Davis, D, Feltell, R, Giraldes, J, Hiscock, S, Kim, S, Kovats, S, Lagu, B, Lewry, K, Loo, A, Lu, Y, Luzzio, M, Maniara, W, Mcmenamin, R, Mortenson, P, Benning, R, O'Reilly, M, Rees, D, Shen, J, Smith, T, Wang, Y, Williams, G, Woolford, A, Wrona, W, Xu, M, Yang, F, Howard, S. | | Deposit date: | 2012-05-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Fragment-Based Discovery of 7-Azabenzimidazoles as Potent, Highly Selective, and Orally Active CDK4/6 Inhibitors.
ACS Med Chem Lett, 3, 2012
|
|
2FDP
 
 | | Crystal structure of beta-secretase complexed with an amino-ethylene inhibitor | | Descriptor: | Beta-secretase 1, N1-((2S,3S,5R)-3-AMINO-6-(4-FLUOROPHENYLAMINO)-5-METHYL-6-OXO-1-PHENYLHEXAN-2-YL)-N3,N3-DIPROPYLISOPHTHALAMIDE | | Authors: | Yang, W, Lu, W, Lu, Y, Zhong, M, Sun, J, Thomas, A.E, Wilkinson, J.M, Fucini, R.V, Lam, M, Randal, M, Shi, X.P, Jacobs, J.W, McDowell, R.S, Gordon, E.M, Ballinger, M.D. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Aminoethylenes: a tetrahedral intermediate isostere yielding potent inhibitors of the aspartyl protease BACE-1.
J.Med.Chem., 49, 2006
|
|
7QXP
 
 | | Proteasome-ZFAND5 Complex Z+B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXX
 
 | | Proteasome-ZFAND5 Complex Z+E state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXU
 
 | | Proteasome-ZFAND5 Complex Z+C state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QY7
 
 | | Proteasome-ZFAND5 Complex Z-A state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QXW
 
 | | Proteasome-ZFAND5 Complex Z+D state | | Descriptor: | 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, 26S protease regulatory subunit 7, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYA
 
 | | Proteasome-ZFAND5 Complex Z-B state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
7QYB
 
 | | Proteasome-ZFAND5 Complex Z-C state | | Descriptor: | 26S protease regulatory subunit 4, 26S protease regulatory subunit 6A, 26S protease regulatory subunit 6B, ... | | Authors: | Zhu, Y, Lu, Y. | | Deposit date: | 2022-01-27 | | Release date: | 2023-02-08 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Molecular mechanism for activation of the 26S proteasome by ZFAND5.
Mol.Cell, 83, 2023
|
|
1HZ3
 
 | | ALZHEIMER'S DISEASE AMYLOID-BETA PEPTIDE (RESIDUES 10-35) | | Descriptor: | A-BETA AMYLOID | | Authors: | Zhang, S, Iwata, K, Lachenmann, M.J, Peng, J.W, Li, S, Stimson, E.R, Lu, Y, Felix, A.M, Maggio, J.E, Lee, J.P. | | Deposit date: | 2001-01-23 | | Release date: | 2001-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Alzheimer's peptide a beta adopts a collapsed coil structure in water.
J.Struct.Biol., 130, 2000
|
|
1LUF
 
 | | Crystal Structure of the MuSK Tyrosine Kinase: Insights into Receptor Autoregulation | | Descriptor: | muscle-specific tyrosine kinase receptor musk | | Authors: | Till, J.H, Becerra, M, Watty, A, Lu, Y, Ma, Y, Neubert, T.A, Burden, S.J, Hubbard, S.R. | | Deposit date: | 2002-05-22 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the MuSK tyrosine kinase: insights into receptor autoregulation.
Structure, 10, 2002
|
|
7WWK
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | Descriptor: | 55A8 light chain, Spike glycoprotein | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7WWJ
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7WWI
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-02-13 | | Release date: | 2023-02-15 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
7XJ9
 
 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab | | Descriptor: | 55A8 heavy chain, 55A8 light chain, 58G6 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab
To Be Published
|
|
7XJ8
 
 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 2 conformation
To Be Published
|
|
7XJ6
 
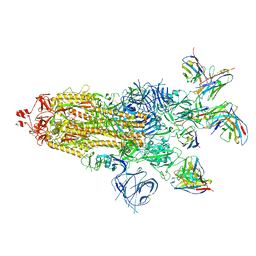 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | Authors: | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | Deposit date: | 2022-04-15 | | Release date: | 2023-04-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab and 58G6 Fab in the class 1 conformation
To Be Published
|
|
7YIV
 
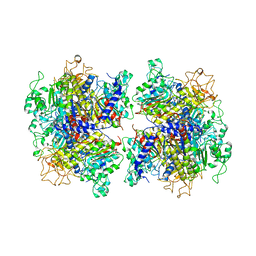 | | The Crystal Structure of Human Tissue Nonspecific Alkaline Phosphatase (ALPL) at Basic pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Cao, Y, Qin, A, Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
7YIW
 
 | | The Crystal Structure of Human Tissue Nonspecific Alkaline Phosphatase (ALPL) at Acidic pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y, Qin, A, Cao, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
4WKX
 
 | |
7YIX
 
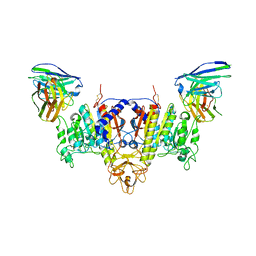 | | The Cryo-EM Structure of Human Tissue Nonspecific Alkaline Phosphatase and Single-Chain Fragment Variable (ScFv) Complex. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Alkaline phosphatase, ... | | Authors: | Yu, Y.T, Yao, D.Q, Zhang, Q, Rao, B, Xia, Y, Lu, Y, Qin, A, Ma, P.X, Cao, Y. | | Deposit date: | 2022-07-18 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The structural pathology for hypophosphatasia caused by malfunctional tissue non-specific alkaline phosphatase.
Nat Commun, 14, 2023
|
|
7SII
 
 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
8K11
 
 | | SID1 transmembrane family member 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K12
 
 | | SID1 transmembrane family member 2 | | Descriptor: | SID1 transmembrane family member 2 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
8K1B
 
 | | SID1 transmembrane family member 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SID1 transmembrane family member 1 | | Authors: | Guo, H, Qi, C, Lu, Y, Yang, H, Zhu, Y, Sun, F, Ji, X. | | Deposit date: | 2023-07-10 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Cryo-EM structures of human SID-1 transmembrane family proteins and implications for their low-pH-dependent RNA transport activity.
Cell Res., 34, 2024
|
|
