7NG3
 
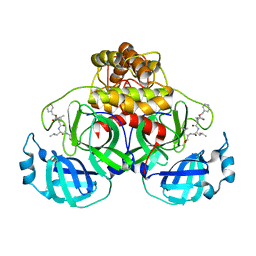 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup P1. | | Descriptor: | 3C-like proteinase, CHLORIDE ION, N-[(benzyloxy)carbonyl]-L-leucyl-N-[(2S)-1-hydroxy-4-methylpentan-2-yl]-L-leucinamide | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7NF5
 
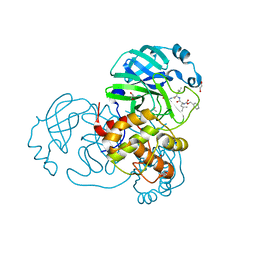 | | Crystal structure of MG-132 covalently bound to the main protease (3CLpro/Mpro) of SARS-CoV-2 in spacegroup C2. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Storici, P. | | Deposit date: | 2021-02-05 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
6RXC
 
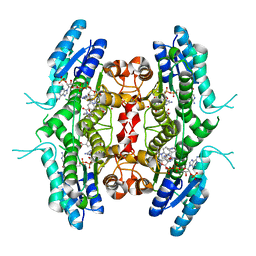 | | Leishmania major pteridine reductase 1 (LmPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1, methyl 1-[4-[[2,4-bis(azanyl)pteridin-6-yl]methyl-(3-oxidanylpropyl)amino]phenyl]carbonylpiperidine-4-carboxylate | | Authors: | Di Pisa, F, Dello Iacono, L, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
6RX5
 
 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with inhibitor 1 (NMT-C0003) | | Descriptor: | ACETATE ION, GLYCEROL, METHYL 1-(4-{[(2,4-DIAMINOPTERIDIN-6-YL)METHYL](METHYL)AMINO}BENZOYL)PIPERIDINE-4-CARBOXYLATE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
6RX0
 
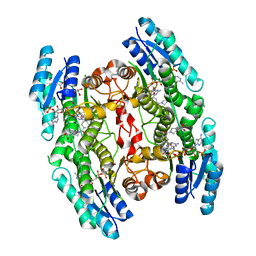 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with inhibitor 3 (NMT-C0013) | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
6RX6
 
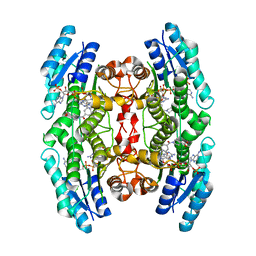 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with inhibitor 4 (NMT-C0026) | | Descriptor: | ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
5L42
 
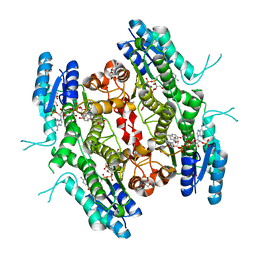 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 3 | | Descriptor: | (2~{R})-2-[3,4-bis(oxidanyl)phenyl]-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
5L4N
 
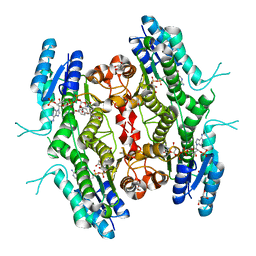 | | Leishmania major Pteridine reductase 1 (PTR1) in complex with compound 1 | | Descriptor: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Dello Iacono, L, Di Pisa, F, Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2016-05-26 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
5K6A
 
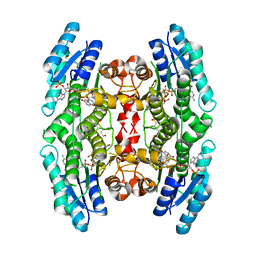 | | Trypanosoma brucei Pteridine reductase 1 (PTR1) in complex with compound 1 | | Descriptor: | (2~{R})-2-(3-hydroxyphenyl)-6-oxidanyl-2,3-dihydrochromen-4-one, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Di Pisa, F, Dello lacono, L, Mangani, S. | | Deposit date: | 2016-05-24 | | Release date: | 2017-03-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Chroman-4-One Derivatives Targeting Pteridine Reductase 1 and Showing Anti-Parasitic Activity.
Molecules, 22, 2017
|
|
6YEK
 
 | | Crystal structure of human NEMO apo form | | Descriptor: | Inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma, isoform CRA_b | | Authors: | Garcia-Pardo, J, Akutsu, M, Busse, P, Skenderovic, A, Maculins, T, Dikic, I. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors by Integrating Protein Engineering and Chemical Screening Platforms.
Cell Chem Biol, 27, 2020
|
|
7ALI
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup P2(1)). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
7ALH
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 at 1.65A resolution (spacegroup C2). | | Descriptor: | 3C-like proteinase | | Authors: | Costanzi, E, Demitri, N, Giabbai, B, Heroux, A, Storici, P. | | Deposit date: | 2020-10-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Biochemical Analysis of the Dual Inhibition of MG-132 against SARS-CoV-2 Main Protease (Mpro/3CLpro) and Human Cathepsin-L.
Int J Mol Sci, 22, 2021
|
|
8P58
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer R. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P86
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM MG-132, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P57
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar X77. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5A
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer R. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P55
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 75 micromolar MG-132. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P87
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 mM X77, from an "old" crystal. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-31 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P56
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar X77. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5C
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P54
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 150 micromolar MG-132. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
8P5B
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 500 micromolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, CHLORIDE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6GD4
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 4c (F188) | | Descriptor: | 2-amino-1,3-benzothiazole-6-carboxamide, ACETATE ION, GLYCEROL, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-21 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCQ
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 2b (F192) | | Descriptor: | 6-(trifluoromethylsulfanyl)-1,3-benzothiazol-2-amine, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
6GCL
 
 | | Trypanosoma brucei PTR1 in complex with inhibitor 3a (F020) | | Descriptor: | 6-methylsulfonyl-1,3-benzothiazol-2-amine, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2018-04-18 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
