5B34
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles in Complex with Iodine-labeled Detergent HAD13a Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | 2,4,6-tris(iodanyl)-5-(octanoylamino)benzene-1,3-dicarboxylic acid, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5B35
 
 | | Serial Femtosecond Crystallography (SFX) of Ground State Bacteriorhodopsin Crystallized from Bicelles Determined Using 7-keV X-ray Free Electron Laser (XFEL) at SACLA | | Descriptor: | (3R,5S,7R,8R,9S,10S,12S,13R,14S,17R)-10,13-dimethyl-17-[(2R)-pentan-2-yl]-2,3,4,5,6,7,8,9,11,12,14,15,16,17-tetradecahydro-1H-cyclopenta[a]phenanthrene-3,7,12-triol, Bacteriorhodopsin, DECANE, ... | | Authors: | Mizohata, E, Nakane, T, Suzuki, M. | | Deposit date: | 2016-02-10 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Membrane protein structure determination by SAD, SIR, or SIRAS phasing in serial femtosecond crystallography using an iododetergent
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
2DQU
 
 | | Crystal form II: high resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
2DQT
 
 | | High resolution crystal structure of the complex of the hydrolytic antibody Fab 6D9 and a transition-state analog | | Descriptor: | IMMUNOGLOBULIN 6D9, [1-(3-DIMETHYLAMINO-PROPYL)-3-ETHYL-UREIDO]-[4-(2,2,2-TRIFLUORO-ACETYLAMINO)-BENZYL]PHOSPHINIC ACID-2-(2,2-DIHYDRO-ACETYLAMINO)-3-HYDROXY-1-(4-NITROPHENYL)-PROPYL ESTER | | Authors: | Kristensen, O, Vassylyev, D.G, Tanaka, F, Ito, N, Morikawa, K, Fujii, I. | | Deposit date: | 2006-05-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thermodynamic and structural basis for transition-state stabilization in antibody-catalyzed hydrolysis
J.Mol.Biol., 369, 2007
|
|
3FKR
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase complex with pyruvate | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION, SODIUM ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-02-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
3FKK
 
 | | Structure of L-2-keto-3-deoxyarabonate dehydratase | | Descriptor: | L-2-keto-3-deoxyarabonate dehydratase, PHOSPHATE ION | | Authors: | Shimada, N, Mikami, B. | | Deposit date: | 2008-12-17 | | Release date: | 2010-01-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of L -2-keto-3-deoxyarabonate dehydratase an enzyme involved in an alternative bacterial pathway of L-arabinose metabolism in complex with pyruvate
To be Published
|
|
3WXT
 
 | |
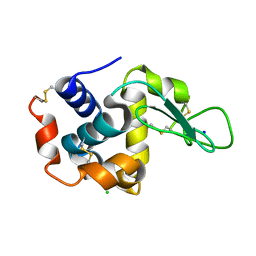
3WUM
 
 | |
3WXU
 
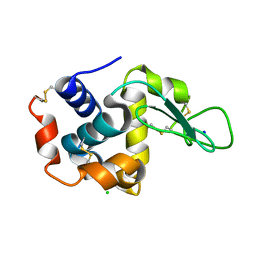 | |
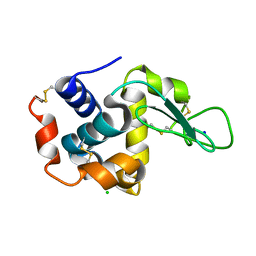
3WUL
 
 | |
3WXS
 
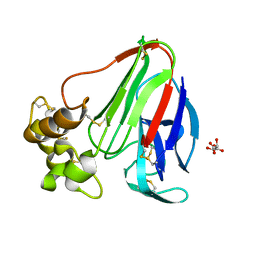 | | Thaumatin structure determined by SPring-8 Angstrom Compact free electron Laser (SACLA) | | Descriptor: | L(+)-TARTARIC ACID, thaumatin I | | Authors: | Masuda, T, Nango, E, Sugahara, M, Mizohata, E, Tanaka, T, Tanaka, R, Suzuki, M, Mikami, B, Iwata, S. | | Deposit date: | 2014-08-07 | | Release date: | 2014-11-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Grease matrix as a versatile carrier of proteins for serial crystallography
Nat. Methods, 12, 2015
|
|
4TKB
 
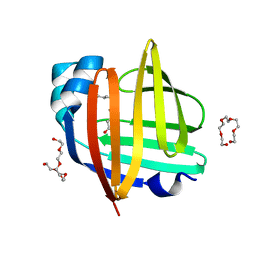 | | The 0.86 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with lauric acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TJZ
 
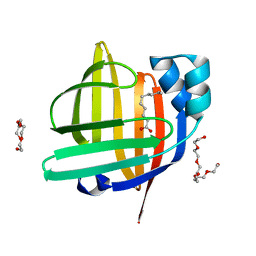 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with capric acid | | Descriptor: | DECANOIC ACID, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKH
 
 | | The 0.93 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with myristic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4TKJ
 
 | | The 0.87 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with palmitic acid | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.87 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type Fatty-Acid-binding protein
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5B5G
 
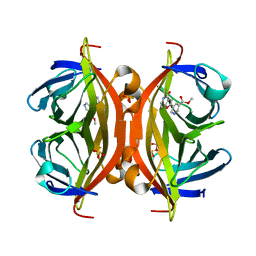 | | Crystal structure of ALiS4-Streptavidin complex | | Descriptor: | SULFITE ION, Streptavidin, methyl 5-(4-oxidanylidene-5~{H}-furo[3,2-c]pyridin-2-yl)pyridine-3-carboxylate | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
5B5F
 
 | | Crystal structure of ALiS3-Streptavidin complex | | Descriptor: | N-methyl-3-(4-oxo-4,5-dihydrofuro[3,2-c]pyridin-2-yl)benzenesulfonamide, Streptavidin | | Authors: | Sugiyama, S, Terai, T, Kakinouchi, K, Fujikake, R, Nagano, T, Urano, Y. | | Deposit date: | 2016-05-04 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Improving the Solubility of Artificial Ligands of Streptavidin to Enable More Practical Reversible Switching of Protein Localization in Cells
Chembiochem, 18, 2017
|
|
3WVM
 
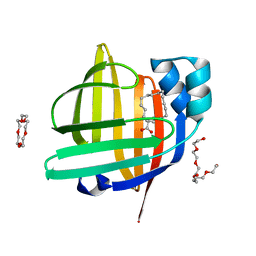 | | The 0.88 angstrom X-ray structure of the human heart fatty acid-binding protein complexed with stearic acid | | Descriptor: | Fatty acid-binding protein, heart, HEXAETHYLENE GLYCOL, ... | | Authors: | Sugiyama, S, Matsuoka, S, Mizohata, E, Matsuoka, D, Ishida, H, Hirose, M, Kakinouchi, K, Hara, T, Matsumura, H, Murakami, S, Inoue, T, Murata, M. | | Deposit date: | 2014-05-25 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (0.88 Å) | | Cite: | Water-mediated recognition of simple alkyl chains by heart-type fatty-acid-binding protein.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
