1YOA
 
 | | Crystal structure of a probable flavoprotein from Thermus thermophilus HB8 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, putative flavoprotein | | Authors: | Imagawa, T, Tsuge, H, Sugimoto, Y, Utsunomiya, H, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-27 | | Release date: | 2005-07-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a flavoenzyme TTHA0420 from Thermus thermophilus HB8
To be published
|
|
1QVC
 
 | | CRYSTAL STRUCTURE ANALYSIS OF SINGLE STRANDED DNA BINDING PROTEIN (SSB) FROM E.COLI | | Descriptor: | SINGLE STRANDED DNA BINDING PROTEIN MONOMER | | Authors: | Matsumoto, T, Morimoto, Y, Shibata, N, Shimamoto, N, Tsukihara, T, Yasuoka, N. | | Deposit date: | 1999-07-07 | | Release date: | 2000-06-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of functional loops and the C-terminal segment of a single-stranded DNA binding protein elucidated by X-Ray structure analysis.
J.Biochem.(Tokyo), 127, 2000
|
|
5B06
 
 | | Lysozyme (denatured by NaOD and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5B07
 
 | | Lysozyme (denatured by DCl and refolded) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
5B05
 
 | | Lysozyme (control experiment) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysozyme C, ... | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2015-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Effective Deuterium Exchange Method for Neutron Crystal Structure Analysis with Unfolding-Refolding Processes
Mol Biotechnol., 58, 2016
|
|
1V7Y
 
 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli at room temperature | | Descriptor: | SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-25 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
3WCQ
 
 | | Crystal structure analysis of Cyanidioschyzon melorae ferredoxin D58N mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Ueno, Y, Matsumoto, T, Yamano, A, Imai, T, Morimoto, Y. | | Deposit date: | 2013-05-31 | | Release date: | 2013-08-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Increasing the electron-transfer ability of Cyanidioschyzon merolae ferredoxin by a one-point mutation - A high resolution and Fe-SAD phasing crystal structure analysis of the Asp58Asn mutant
Biochem.Biophys.Res.Commun., 436, 2013
|
|
3W69
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | (5R,6S)-2-[((2S,5R)-2-{[(3R)-4-acetyl-3-methylpiperazin-1-yl]carbonyl}-5-ethylpyrrolidin-1-yl)carbonyl]-5,6-bis(4-chlorophenyl)-3-isopropyl-6-methyl-5,6-dihydroimidazo[2,1-b][1,3]thiazole, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2013-02-12 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and evaluation of novel orally active p53-MDM2 interaction inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
2DH6
 
 | | Crystal structure of E. coli Apo-TrpB | | Descriptor: | SULFATE ION, tryptophan synthase beta subunit | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yasuoka, N, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
2DH5
 
 | | Crystal structure of E. coli Holo-TrpB | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Nishio, K, Morimoto, Y, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large conformational changes in the Escherichia coli tryptophan synthase beta(2) subunit upon pyridoxal 5'-phosphate binding
Febs J., 277, 2010
|
|
6K8G
 
 | | H/D exchanged Hen egg-white lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Kita, A, Morimoto, Y. | | Deposit date: | 2019-06-11 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2 Å), X-RAY DIFFRACTION | | Cite: | Hydrogen/deuterium exchange behavior in tetragonal hen egg-white lysozyme crystals affected by solution state.
J.Appl.Crystallogr., 53, 2020
|
|
3VZV
 
 | | Crystal structure of human mdm2 with a dihydroimidazothiazole inhibitor | | Descriptor: | 1-{[(5R,6S)-5,6-bis(4-chlorophenyl)-6-methyl-3-(propan-2-yl)-5,6-dihydroimidazo[2,1-b][1,3]thiazol-2-yl]carbonyl}-N,N-dimethyl-L-prolinamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Shimizu, H, Katakura, S, Miyazaki, M, Naito, H, Sugimoto, Y, Kawato, H, Okayama, T, Soga, T. | | Deposit date: | 2012-10-16 | | Release date: | 2013-02-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lead optimization of novel p53-MDM2 interaction inhibitors possessing dihydroimidazothiazole scaffold
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3WMQ
 
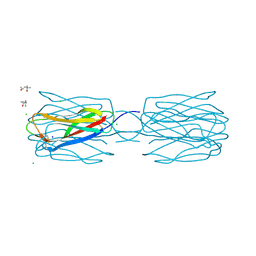 | | Crystal structure of the complex between SLL-2 and GalNAc. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WMP
 
 | | Crystal structure of SLL-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kita, A, Jimbo, M, Sakai, R, Morimoto, Y, Miki, K. | | Deposit date: | 2013-11-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a symbiosis-related lectin from octocoral.
Glycobiology, 25, 2015
|
|
3WKT
 
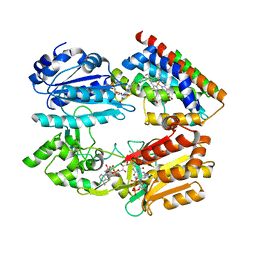 | | Complex structure of an open form of NADPH-cytochrome P450 reductase and heme oxygenase-1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Heme oxygenase 1, ... | | Authors: | Sugishima, M, Sato, H, Higashimoto, Y, Harada, J, Wada, K, Fukuyama, K, Noguchi, M. | | Deposit date: | 2013-10-31 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis for the electron transfer from an open form of NADPH-cytochrome P450 oxidoreductase to heme oxygenase.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7FGV
 
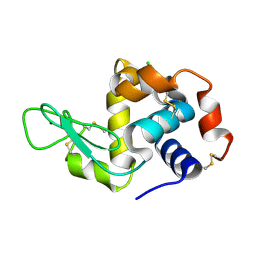 | |
7FG8
 
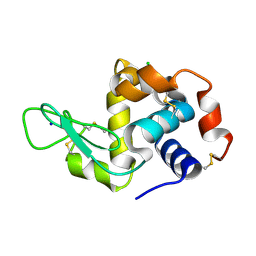 | |
7FGU
 
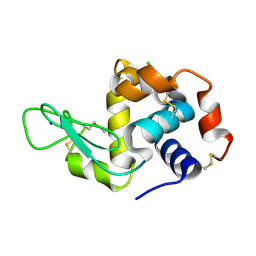 | |
2ZBW
 
 | | Crystal structure of thioredoxin reductase-like protein from Thermus thermophilus HB8 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase | | Authors: | Ebihara, A, Manzoku, M, Fujimoto, Y, Kitamura, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thioredoxin reductase-like protein from Thermus thermophilus HB8
To be Published
|
|
2Z6W
 
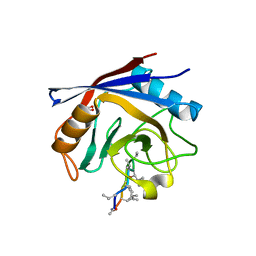 | | Crystal structure of human cyclophilin D in complex with cyclosporin A | | Descriptor: | CITRIC ACID, CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE | | Authors: | Kajitani, K, Fujihashi, M, Kobayashi, Y, Shimizu, S, Tsujimoto, Y, Miki, K. | | Deposit date: | 2007-08-09 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Crystal Structure of Human Cyclophilin D in Complex with its Inhibitor, Cyclosporin a at 0.96-A Resolution.
Proteins, 70, 2008
|
|
2YWN
 
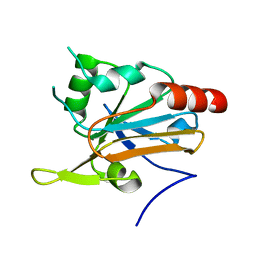 | | Crystal structure of peroxiredoxin-like protein from Sulfolobus tokodaii | | Descriptor: | Peroxiredoxin-like protein | | Authors: | Ebihara, A, Manzoku, M, Fujimoto, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-20 | | Release date: | 2007-10-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of peroxiredoxin-like protein from Sulfolobus tokodaii
To be Published
|
|
2YZH
 
 | | Crystal structure of peroxiredoxin-like protein from Aquifex aeolicus | | Descriptor: | Probable thiol peroxidase, SULFATE ION | | Authors: | Ebihara, A, Manzoku, M, Fujimoto, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-05 | | Release date: | 2007-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of peroxiredoxin-like protein from Aquifex aeolicus
to be published
|
|
2ZBV
 
 | | Crystal structure of uncharacterized conserved protein from Thermotoga maritima | | Descriptor: | ADENOSINE, Uncharacterized conserved protein | | Authors: | Ebihara, A, Fujimoto, Y, Kagawa, W, Fujikawa, N, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Thermotoga maritima
To be Published
|
|
2ZK1
 
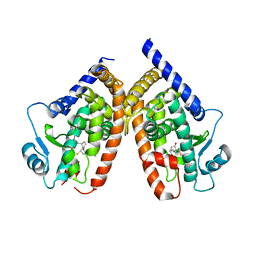 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with 15-deoxy-delta12,14-prostaglandin J2 | | Descriptor: | (5E,14E)-11-oxoprosta-5,9,12,14-tetraen-1-oic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
2ZK4
 
 | | Human peroxisome proliferator-activated receptor gamma ligand binding domain complexed with 15-oxo-eicosatetraenoic acid | | Descriptor: | (5E,8E,11Z,13E)-15-oxoicosa-5,8,11,13-tetraenoic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Waku, T, Shiraki, T, Oyama, T, Fujimoto, Y, Morikawa, K. | | Deposit date: | 2008-03-12 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural insight into PPARgamma activation through covalent modification with endogenous fatty acids
J.Mol.Biol., 385, 2009
|
|
