6FZD
 
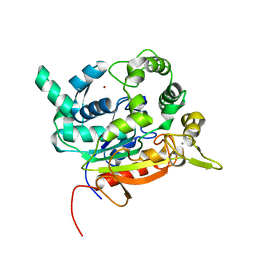 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 variant L184F/A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ1
 
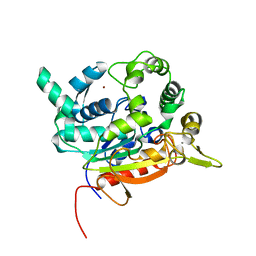 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-13 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ9
 
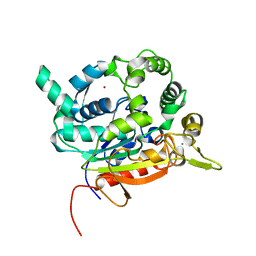 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant A187F/L360F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.2463 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
6FZ7
 
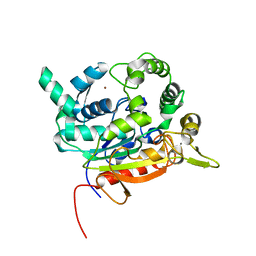 | | Crystal Structure of lipase from Geobacillus stearothermophilus T6 methanol stable variant L184F | | Descriptor: | CALCIUM ION, Lipase, ZINC ION | | Authors: | Gihaz, S, Kanteev, M, Pazy, Y, Fishman, A. | | Deposit date: | 2018-03-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.736 Å) | | Cite: | Filling the Void: Introducing Aromatic Interactions into Solvent Tunnels To Enhance Lipase Stability in Methanol.
Appl.Environ.Microbiol., 84, 2018
|
|
1AFT
 
 | | SMALL SUBUNIT C-TERMINAL INHIBITORY PEPTIDE OF MOUSE RIBONUCLEOTIDE REDUCTASE AS BOUND TO THE LARGE SUBUNIT, NMR, 26 STRUCTURES | | Descriptor: | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE | | Authors: | Laub, P.B, Fisher, A.L, Furst, G.T, Barwis, B.A, Hamann, C.S, Cooperman, B.S. | | Deposit date: | 1997-03-13 | | Release date: | 1997-05-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of an inhibitory R2 C-terminal peptide bound to mouse ribonucleotide reductase R1 subunit.
Nat.Struct.Biol., 2, 1995
|
|
1WB9
 
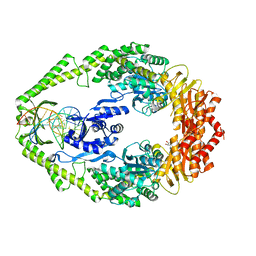 | | Crystal Structure of E. coli DNA Mismatch Repair enzyme MutS, E38T mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBB
 
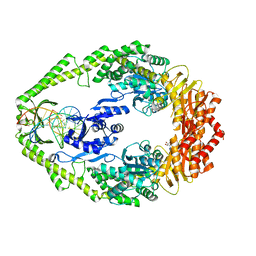 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38A mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
1WBD
 
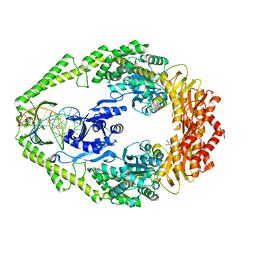 | | Crystal structure of E. coli DNA mismatch repair enzyme MutS, E38Q mutant, in complex with a G.T mismatch | | Descriptor: | 5'-D(*AP*CP*TP*GP*GP*TP*GP*CP*TP*TP *GP*GP*CP*AP*GP*CP*T)-3', 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP *CP*AP*CP*CP*AP*GP*TP*G)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Natrajan, G, Georgijevic, D, Lebbink, J.H.G, Winterwerp, H.H.K, de Wind, N, Sixma, T.K. | | Deposit date: | 2004-10-31 | | Release date: | 2006-01-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dual Role of Muts Glutamate 38 in DNA Mismatch Discrimination and in the Authorization of Repair.
Embo J., 25, 2006
|
|
4PG9
 
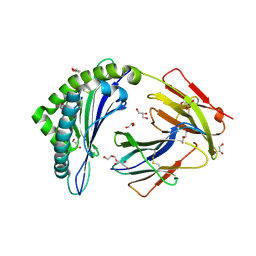 | | MHC Class I in complex with Sendai virus nucleoprotein peptide FAPGNYPAL | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Celie, P, Joosten, R.P, Perrakis, A, Neefjes, J. | | Deposit date: | 2014-05-01 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The first step of peptide selection in antigen presentation by MHC class I molecules.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4PGE
 
 | | MHC Class I in complex with modified Sendai virus nucleoprotein peptide FAPGNYPAW | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Celie, P, Joosten, R.P, Perrakis, A, Neefjes, J. | | Deposit date: | 2014-05-01 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The first step of peptide selection in antigen presentation by MHC class I molecules.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4PGD
 
 | | MHC Class I in complex with modified Sendai virus nucleoprotein peptide FAPGNYPAF | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Celie, P, Joosten, R.P, Perrakis, A, Neefjes, J. | | Deposit date: | 2014-05-01 | | Release date: | 2015-01-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The first step of peptide selection in antigen presentation by MHC class I molecules.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4PGB
 
 | | MHC Class I in complex with modified Sendai virus nucleoprotein peptide FAPGNWPAL | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Celie, P.H.N, Joosten, R.P, Perrakis, A, Neefjes, J. | | Deposit date: | 2014-05-01 | | Release date: | 2015-01-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The first step of peptide selection in antigen presentation by MHC class I molecules.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5C9N
 
 | | Crystal structure of GEMC1 coiled-coil domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Geminin coiled-coil domain-containing protein 1 | | Authors: | Caillat, C, Perrakis, A. | | Deposit date: | 2015-06-28 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled-coil proteins.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5L8W
 
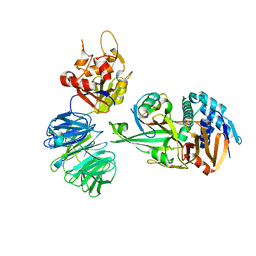 | | Structure of USP12-UB-PRG/UAF1 | | Descriptor: | GLYCEROL, Polyubiquitin-B, Ubiquitin carboxyl-terminal hydrolase 12, ... | | Authors: | Dharadhar, S, Sixma, T. | | Deposit date: | 2016-06-08 | | Release date: | 2016-09-28 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | A conserved two-step binding for the UAF1 regulator to the USP12 deubiquitinating enzyme.
J.Struct.Biol., 196, 2016
|
|
3P63
 
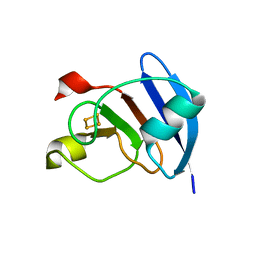 | | Structure of M. laminosus Ferredoxin with a shorter L1,2 loop | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Ferredoxin | | Authors: | Livnah, O, Nechushtai, R, Eisenberg-Domovich, Y, Michaeli, D. | | Deposit date: | 2010-10-11 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allostery in the ferredoxin protein motif does not involve a conformational switch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6FPV
 
 | | A llama-derived JBP1-targeting nanobody | | Descriptor: | GLYCEROL, Nanobody | | Authors: | van Beusekom, B, Adamopoulos, A, Heidebrecht, T, Joosten, R.P, Perrakis, A. | | Deposit date: | 2018-02-12 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Characterization and structure determination of a llama-derived nanobody targeting the J-base binding protein 1.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
3K0S
 
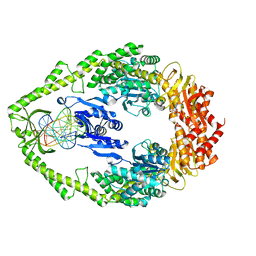 | | Crystal structure of E.coli DNA mismatch repair protein MutS, D693N mutant, in complex with GT mismatched DNA | | Descriptor: | 5'-D(*AP*GP*CP*TP*GP*CP*CP*AP*GP*GP*CP*AP*CP*CP*AP*GP*TP*GP*TP*CP*AP*GP*CP*GP*TP*CP*CP*TP*AP*T)-3', 5'-D(*AP*TP*AP*GP*GP*AP*CP*GP*CP*TP*GP*AP*C*AP*CP*T*GP*GP*TP*GP*CP*TP*TP*GP*GP*CP*AP*GP*CP*T)-3', ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Reumer, G.A, Winterwerp, H.H.K, Sixma, T.K. | | Deposit date: | 2009-09-25 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Magnesium coordination controls the molecular switch function of DNA mismatch repair protein MutS.
J.Biol.Chem., 285, 2010
|
|
8AG6
 
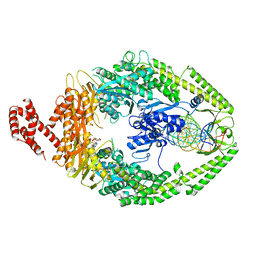 | |
4Z2P
 
 | |
4Z27
 
 | |
4Z2V
 
 | |
4Z2O
 
 | |
4Z28
 
 | | Crystal structure of short hoefavidin biotin complex | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Avidin family, BIOTIN | | Authors: | Livnah, O, Avraham, O. | | Deposit date: | 2015-03-29 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Hoefavidin: A dimeric bacterial avidin with a C-terminal binding tail.
J.Struct.Biol., 191, 2015
|
|
4Z6J
 
 | |
6YJP
 
 | | Crystal structure of a complex between glycosylated NKp30 and its deglycosylated tumour ligand B7-H6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Natural cytotoxicity triggering receptor 3, Natural cytotoxicity triggering receptor 3 ligand 1 | | Authors: | Skalova, T, Dohnalek, J, Skorepa, O, Kalouskova, B, Pazicky, S, Blaha, J, Vanek, O. | | Deposit date: | 2020-04-04 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Natural Killer Cell Activation Receptor NKp30 Oligomerization Depends on Its N -Glycosylation.
Cancers (Basel), 12, 2020
|
|
